Gene
KWMTBOMO12645
Pre Gene Modal
BGIBMGA001567
Annotation
PREDICTED:_actin-binding_Rho-activating_protein-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.431
Sequence
CDS
ATGAGCGGCAATTTCCTAACGATACCCGGAGATGATGCTATTCAGAGACGATCAAGGAGCTCGTCGCTTACGGACTTGATATCCGCCTTCGACACAAAAGCACAGGCTCACACCGAGCTTCAGAAAGTAACAGCCTTCAGCGGACAATTCGACAAGAACCACAAGCCTACATTCTCAAAAGACGAATATGGAAAACCTATACCGGGTTCGATGACCGAATACAGAGGCGCAGAAGCACAAGCCAGAGTATGTACAGAAATGAAGGAGCTTTGTGAAATCATCAACGAATACGGAAAGACCCCATGCAAGAGTTTATTACCGCCTGAATTACAAGACGCCACCAAAGTTATATCATTTGGAGAATTATTTACTATATACACGGTGATATCGGACAAACTGGTTGGGATACTTCTGAGAACGCGGAAACATCAGCTAACGTTCTTCGAAGGAGAAGTATTGTTTCAGAAACGTGACGATGATGTTCCGGTTTTCCTGATGCAACCCATCGACGATATAAGAACGTATTGCAACAACAAAGCGCTTCAGGCCAGAAGATCGGTGTCGCCTATGCCCAATTAA
Protein
MSGNFLTIPGDDAIQRRSRSSSLTDLISAFDTKAQAHTELQKVTAFSGQFDKNHKPTFSKDEYGKPIPGSMTEYRGAEAQARVCTEMKELCEIINEYGKTPCKSLLPPELQDATKVISFGELFTIYTVISDKLVGILLRTRKHQLTFFEGEVLFQKRDDDVPVFLMQPIDDIRTYCNNKALQARRSVSPMPN
Summary
Uniprot
A0A194QCJ3
A0A212EI14
A0A1E1WUI5
A0A437BUY2
A0A194RDW0
A0A0T6B3Q5
+ More
A0A1Y1KE37 T1PG31 B0XHU2 A0A3B0K092 A0A1B0ALU1 A0A1I8NFN8 B4I9Q5 A0A1A9X7Z0 O46052 B4L6S8 B3P8X2 A0A1A9UU79 B3MR31 B4Q1A2 A0A0M4EXD9 B4GXS2 A0A1I8Q0U3 A0A1Q3FYL1 Q29HF9 A0A1A9W290 B4NC46 A0A1B0FDR1 A0A1W4VEH6 A0A1B0AJZ2 B4M7R9 A0A0C9QJS9 A0A0Q5ST70 B4R3F0 M9PDL0 A0A0P9A9P4 A0A0R3P078 A0A1I8Q0X6 A0A0L0BZY5 A0A0R1E9T0 A0A1W7R4S8 A0A023EME2 A0A0Q9WU36 A0A1W4VR78 B4JMM8 D6WN35 W8C290 W8BFX2 A0A034VT38 A0A182WS82 Q380I4 A0A034VNU8 A0A182FRI3 A0A0C9RYU8 T1E9U9 A0A182IQA8 A0A0K8UV05 A0A310SHC3 A0A154PLS5 A0A2A3ECJ6 A0A088AD70 A0A0A1XT20 K7J3V2 A0A182UUL4 U5EPQ8 A0A182JSQ3 A0A0L7R9Y3 A0A158NSV7 A0A182QGV7 A0A195CST8 A0A182YAI1 J3JVY7 A0A182R4Y5 A0A182MIF5 F5HMN1 A0A182VWF8 A0A182PGX7 A0A151WYT1 A0A195DPW0 A0A182L9F1 A0A1B6CIQ5 A0A182UD78 A0A182HL31 A0A151I0J9 A0A087TGV2 A0A3L8DUE4 A0A0N0U679 A0A1J1HRN5 A0A182NQD6 V5GHL5 E2AFT8 F4WZ62 A0A2S2QX27 A0A1B6GIJ3 A0A026X1Y5 A0A0A9WQF7 A0A195EXP1 A0A1J1HQ05
A0A1Y1KE37 T1PG31 B0XHU2 A0A3B0K092 A0A1B0ALU1 A0A1I8NFN8 B4I9Q5 A0A1A9X7Z0 O46052 B4L6S8 B3P8X2 A0A1A9UU79 B3MR31 B4Q1A2 A0A0M4EXD9 B4GXS2 A0A1I8Q0U3 A0A1Q3FYL1 Q29HF9 A0A1A9W290 B4NC46 A0A1B0FDR1 A0A1W4VEH6 A0A1B0AJZ2 B4M7R9 A0A0C9QJS9 A0A0Q5ST70 B4R3F0 M9PDL0 A0A0P9A9P4 A0A0R3P078 A0A1I8Q0X6 A0A0L0BZY5 A0A0R1E9T0 A0A1W7R4S8 A0A023EME2 A0A0Q9WU36 A0A1W4VR78 B4JMM8 D6WN35 W8C290 W8BFX2 A0A034VT38 A0A182WS82 Q380I4 A0A034VNU8 A0A182FRI3 A0A0C9RYU8 T1E9U9 A0A182IQA8 A0A0K8UV05 A0A310SHC3 A0A154PLS5 A0A2A3ECJ6 A0A088AD70 A0A0A1XT20 K7J3V2 A0A182UUL4 U5EPQ8 A0A182JSQ3 A0A0L7R9Y3 A0A158NSV7 A0A182QGV7 A0A195CST8 A0A182YAI1 J3JVY7 A0A182R4Y5 A0A182MIF5 F5HMN1 A0A182VWF8 A0A182PGX7 A0A151WYT1 A0A195DPW0 A0A182L9F1 A0A1B6CIQ5 A0A182UD78 A0A182HL31 A0A151I0J9 A0A087TGV2 A0A3L8DUE4 A0A0N0U679 A0A1J1HRN5 A0A182NQD6 V5GHL5 E2AFT8 F4WZ62 A0A2S2QX27 A0A1B6GIJ3 A0A026X1Y5 A0A0A9WQF7 A0A195EXP1 A0A1J1HQ05
Pubmed
26354079
22118469
28004739
25315136
17994087
10731132
+ More
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 26108605 24945155 18362917 19820115 24495485 25348373 12364791 14747013 17210077 25830018 20075255 21347285 25244985 22516182 23537049 20966253 30249741 20798317 21719571 24508170 25401762 26823975
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 26108605 24945155 18362917 19820115 24495485 25348373 12364791 14747013 17210077 25830018 20075255 21347285 25244985 22516182 23537049 20966253 30249741 20798317 21719571 24508170 25401762 26823975
EMBL
KQ459193
KPJ03139.1
AGBW02014739
OWR41117.1
GDQN01001199
GDQN01000345
+ More
JAT89855.1 JAT90709.1 RSAL01000006 RVE54278.1 KQ460325 KPJ15787.1 LJIG01016140 KRT81495.1 GEZM01086144 JAV59742.1 KA647100 AFP61729.1 DS233206 EDS28665.1 OUUW01000003 SPP78421.1 JXJN01000141 CH480825 EDW43936.1 AE014298 AY060993 AL009194 AAF45738.3 AAL28541.1 AGB95023.1 CAA15697.1 CH933812 EDW06074.2 CH954183 EDV45577.2 CH902622 EDV34236.1 CM000162 EDX01409.1 CP012528 ALC48380.1 CH479196 EDW27549.1 GFDL01002412 JAV32633.1 CH379064 EAL31799.2 CH964239 EDW82405.2 CCAG010016762 CH940653 EDW62836.1 GBYB01000852 JAG70619.1 KQS26031.1 CM000366 EDX16902.1 AGB95022.1 KPU75038.1 KRT06572.1 JRES01001093 KNC25627.1 KRK06140.1 GEHC01001491 JAV46154.1 GAPW01004114 JAC09484.1 KRF99611.1 CH916371 EDV91971.1 KQ971343 EFA04345.1 GAMC01010706 JAB95849.1 GAMC01010707 JAB95848.1 GAKP01013952 JAC45000.1 AAAB01008987 EAL38580.4 GAKP01013951 JAC45001.1 GBYB01014515 JAG84282.1 GAMD01000982 JAB00609.1 GDHF01021903 JAI30411.1 KQ759903 OAD62000.1 KQ434943 KZC12238.1 KZ288285 PBC29503.1 GBXI01000096 JAD14196.1 GANO01004588 JAB55283.1 KQ414620 KOC67682.1 ADTU01025214 AXCN02001526 KQ977305 KYN03768.1 APGK01051402 BT127405 KB741190 KB632094 AEE62367.1 ENN73012.1 ERL88707.1 AXCM01000785 EGK97553.1 KQ982649 KYQ52946.1 KQ980653 KYN14847.1 GEDC01023942 JAS13356.1 APCN01000879 KQ976619 KYM79140.1 KK115167 KFM64341.1 QOIP01000004 RLU23912.1 KQ435755 KOX76051.1 CVRI01000015 CRK90052.1 GALX01007344 JAB61122.1 GL439158 EFN67687.1 GL888470 EGI60513.1 GGMS01013010 MBY82213.1 GECZ01007646 GECZ01004379 GECZ01000947 JAS62123.1 JAS65390.1 JAS68822.1 KK107054 EZA61404.1 GBHO01036529 GBRD01014397 GDHC01010840 JAG07075.1 JAG51429.1 JAQ07789.1 KQ981923 KYN32916.1 CRK90051.1
JAT89855.1 JAT90709.1 RSAL01000006 RVE54278.1 KQ460325 KPJ15787.1 LJIG01016140 KRT81495.1 GEZM01086144 JAV59742.1 KA647100 AFP61729.1 DS233206 EDS28665.1 OUUW01000003 SPP78421.1 JXJN01000141 CH480825 EDW43936.1 AE014298 AY060993 AL009194 AAF45738.3 AAL28541.1 AGB95023.1 CAA15697.1 CH933812 EDW06074.2 CH954183 EDV45577.2 CH902622 EDV34236.1 CM000162 EDX01409.1 CP012528 ALC48380.1 CH479196 EDW27549.1 GFDL01002412 JAV32633.1 CH379064 EAL31799.2 CH964239 EDW82405.2 CCAG010016762 CH940653 EDW62836.1 GBYB01000852 JAG70619.1 KQS26031.1 CM000366 EDX16902.1 AGB95022.1 KPU75038.1 KRT06572.1 JRES01001093 KNC25627.1 KRK06140.1 GEHC01001491 JAV46154.1 GAPW01004114 JAC09484.1 KRF99611.1 CH916371 EDV91971.1 KQ971343 EFA04345.1 GAMC01010706 JAB95849.1 GAMC01010707 JAB95848.1 GAKP01013952 JAC45000.1 AAAB01008987 EAL38580.4 GAKP01013951 JAC45001.1 GBYB01014515 JAG84282.1 GAMD01000982 JAB00609.1 GDHF01021903 JAI30411.1 KQ759903 OAD62000.1 KQ434943 KZC12238.1 KZ288285 PBC29503.1 GBXI01000096 JAD14196.1 GANO01004588 JAB55283.1 KQ414620 KOC67682.1 ADTU01025214 AXCN02001526 KQ977305 KYN03768.1 APGK01051402 BT127405 KB741190 KB632094 AEE62367.1 ENN73012.1 ERL88707.1 AXCM01000785 EGK97553.1 KQ982649 KYQ52946.1 KQ980653 KYN14847.1 GEDC01023942 JAS13356.1 APCN01000879 KQ976619 KYM79140.1 KK115167 KFM64341.1 QOIP01000004 RLU23912.1 KQ435755 KOX76051.1 CVRI01000015 CRK90052.1 GALX01007344 JAB61122.1 GL439158 EFN67687.1 GL888470 EGI60513.1 GGMS01013010 MBY82213.1 GECZ01007646 GECZ01004379 GECZ01000947 JAS62123.1 JAS65390.1 JAS68822.1 KK107054 EZA61404.1 GBHO01036529 GBRD01014397 GDHC01010840 JAG07075.1 JAG51429.1 JAQ07789.1 KQ981923 KYN32916.1 CRK90051.1
Proteomes
UP000053268
UP000007151
UP000283053
UP000053240
UP000095301
UP000002320
+ More
UP000268350 UP000092460 UP000001292 UP000092443 UP000000803 UP000009192 UP000008711 UP000078200 UP000007801 UP000002282 UP000092553 UP000008744 UP000095300 UP000001819 UP000091820 UP000007798 UP000092444 UP000192221 UP000092445 UP000008792 UP000000304 UP000037069 UP000001070 UP000007266 UP000076407 UP000007062 UP000069272 UP000075880 UP000076502 UP000242457 UP000005203 UP000002358 UP000075903 UP000075881 UP000053825 UP000005205 UP000075886 UP000078542 UP000076408 UP000019118 UP000030742 UP000075900 UP000075883 UP000075920 UP000075885 UP000075809 UP000078492 UP000075882 UP000075902 UP000075840 UP000078540 UP000054359 UP000279307 UP000053105 UP000183832 UP000075884 UP000000311 UP000007755 UP000053097 UP000078541
UP000268350 UP000092460 UP000001292 UP000092443 UP000000803 UP000009192 UP000008711 UP000078200 UP000007801 UP000002282 UP000092553 UP000008744 UP000095300 UP000001819 UP000091820 UP000007798 UP000092444 UP000192221 UP000092445 UP000008792 UP000000304 UP000037069 UP000001070 UP000007266 UP000076407 UP000007062 UP000069272 UP000075880 UP000076502 UP000242457 UP000005203 UP000002358 UP000075903 UP000075881 UP000053825 UP000005205 UP000075886 UP000078542 UP000076408 UP000019118 UP000030742 UP000075900 UP000075883 UP000075920 UP000075885 UP000075809 UP000078492 UP000075882 UP000075902 UP000075840 UP000078540 UP000054359 UP000279307 UP000053105 UP000183832 UP000075884 UP000000311 UP000007755 UP000053097 UP000078541
Pfam
PF14705 Costars
Gene 3D
ProteinModelPortal
A0A194QCJ3
A0A212EI14
A0A1E1WUI5
A0A437BUY2
A0A194RDW0
A0A0T6B3Q5
+ More
A0A1Y1KE37 T1PG31 B0XHU2 A0A3B0K092 A0A1B0ALU1 A0A1I8NFN8 B4I9Q5 A0A1A9X7Z0 O46052 B4L6S8 B3P8X2 A0A1A9UU79 B3MR31 B4Q1A2 A0A0M4EXD9 B4GXS2 A0A1I8Q0U3 A0A1Q3FYL1 Q29HF9 A0A1A9W290 B4NC46 A0A1B0FDR1 A0A1W4VEH6 A0A1B0AJZ2 B4M7R9 A0A0C9QJS9 A0A0Q5ST70 B4R3F0 M9PDL0 A0A0P9A9P4 A0A0R3P078 A0A1I8Q0X6 A0A0L0BZY5 A0A0R1E9T0 A0A1W7R4S8 A0A023EME2 A0A0Q9WU36 A0A1W4VR78 B4JMM8 D6WN35 W8C290 W8BFX2 A0A034VT38 A0A182WS82 Q380I4 A0A034VNU8 A0A182FRI3 A0A0C9RYU8 T1E9U9 A0A182IQA8 A0A0K8UV05 A0A310SHC3 A0A154PLS5 A0A2A3ECJ6 A0A088AD70 A0A0A1XT20 K7J3V2 A0A182UUL4 U5EPQ8 A0A182JSQ3 A0A0L7R9Y3 A0A158NSV7 A0A182QGV7 A0A195CST8 A0A182YAI1 J3JVY7 A0A182R4Y5 A0A182MIF5 F5HMN1 A0A182VWF8 A0A182PGX7 A0A151WYT1 A0A195DPW0 A0A182L9F1 A0A1B6CIQ5 A0A182UD78 A0A182HL31 A0A151I0J9 A0A087TGV2 A0A3L8DUE4 A0A0N0U679 A0A1J1HRN5 A0A182NQD6 V5GHL5 E2AFT8 F4WZ62 A0A2S2QX27 A0A1B6GIJ3 A0A026X1Y5 A0A0A9WQF7 A0A195EXP1 A0A1J1HQ05
A0A1Y1KE37 T1PG31 B0XHU2 A0A3B0K092 A0A1B0ALU1 A0A1I8NFN8 B4I9Q5 A0A1A9X7Z0 O46052 B4L6S8 B3P8X2 A0A1A9UU79 B3MR31 B4Q1A2 A0A0M4EXD9 B4GXS2 A0A1I8Q0U3 A0A1Q3FYL1 Q29HF9 A0A1A9W290 B4NC46 A0A1B0FDR1 A0A1W4VEH6 A0A1B0AJZ2 B4M7R9 A0A0C9QJS9 A0A0Q5ST70 B4R3F0 M9PDL0 A0A0P9A9P4 A0A0R3P078 A0A1I8Q0X6 A0A0L0BZY5 A0A0R1E9T0 A0A1W7R4S8 A0A023EME2 A0A0Q9WU36 A0A1W4VR78 B4JMM8 D6WN35 W8C290 W8BFX2 A0A034VT38 A0A182WS82 Q380I4 A0A034VNU8 A0A182FRI3 A0A0C9RYU8 T1E9U9 A0A182IQA8 A0A0K8UV05 A0A310SHC3 A0A154PLS5 A0A2A3ECJ6 A0A088AD70 A0A0A1XT20 K7J3V2 A0A182UUL4 U5EPQ8 A0A182JSQ3 A0A0L7R9Y3 A0A158NSV7 A0A182QGV7 A0A195CST8 A0A182YAI1 J3JVY7 A0A182R4Y5 A0A182MIF5 F5HMN1 A0A182VWF8 A0A182PGX7 A0A151WYT1 A0A195DPW0 A0A182L9F1 A0A1B6CIQ5 A0A182UD78 A0A182HL31 A0A151I0J9 A0A087TGV2 A0A3L8DUE4 A0A0N0U679 A0A1J1HRN5 A0A182NQD6 V5GHL5 E2AFT8 F4WZ62 A0A2S2QX27 A0A1B6GIJ3 A0A026X1Y5 A0A0A9WQF7 A0A195EXP1 A0A1J1HQ05
PDB
2KRH
E-value=1.14523e-12,
Score=173
Ontologies
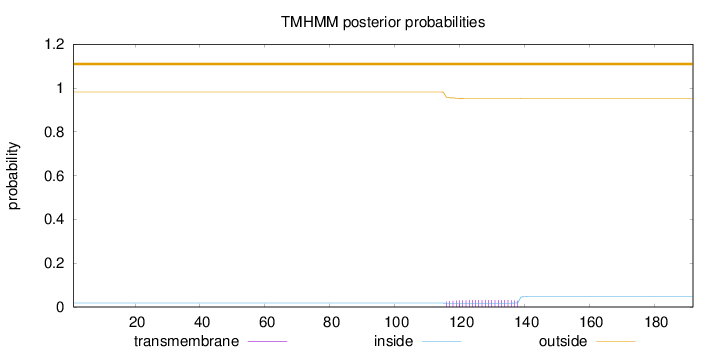
Topology
Length:
192
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.69883
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01777
outside
1 - 192
Population Genetic Test Statistics
Pi
247.459731
Theta
180.7691
Tajima's D
0.965984
CLR
0.388173
CSRT
0.648717564121794
Interpretation
Uncertain
