Gene
KWMTBOMO12644
Annotation
PREDICTED:_protein_unc-13_homolog_A_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.404
Sequence
CDS
ATGTCTACGCTATTAGCTAACATTAATGCCTACTACGCGCACACGACAGCGTCATCGGCTGTGTCTGCATCAGATAGATTCGCGGCTTCAAACTTTGGGAAAGAGAAATTCGTGAAACTGCTCGATCAACTTCACAACTCTCTCAGAATCGATCTCTCGATGTATCGAAACAACTTCCCCGCTTCGAGTGCTGAGAAACTCATGGACCTGAAGTCTACTGTTGACCTGTTGACCAGCATCACTTTCTTTAGGATGAAGGTCCAAGAGTTGAGCAGTCCTCCCCGAGCTAGCACAGTGGTTAAAGACTGCGTGAAAGCCTGTTTGAGGTCAACATACCAGTTCTTATTCGAAAATTGCTACGAGCTTTACAATAGAGAATTTCAGGTAGACCCCAATGAAGCAAAGCGTGACCCAGCTGACCACGGACCACAGTTAGACTCTGTCGATTTCTGGCACAAATTAATAGCTCTCATAGTGTCTGTTATAGAAGAAGACCGAAATAGCTACGCTCCCGTACTGAACCAGTTCCCGCAGGAGCTCAATATTGGACAGCTGTCCGCAGCCACAATGTGGTCGTTATTCGCAGTGGATATGAAGTACGCTCTGGAGGAACATGAACAACACAGGCTCTGCAAGTCTTCCGCCTACATGAATCTCCACTTCAAAGTAAAATGGCTGCATACGAACTACGTGAAAGACGTCCCGCCGTACTGTGGCGCTGTGCCGGAGTATCCGGCGTGGTTCGAACCCTTCGTGATGCAGTGGCTGAATGAAAACGACGACGTGTCCTTAGAATATTTGAACGGAGCATTCAACAGAGATAAACGTGATGGGTTCCAAAAGTCCAGCGAGCACGCGCTCTTCAGCAACTCAGTGGTCGACGTGTTCACGCAACTGACGCAATGCTTCGACGTGGTCTCGAAGCTGGAGTGTCCCGACCCTGAAATATGGAAACGGTACATGAAACGTTTCGCCAAGACCATAGTGAAGGTGCTGGTCGCGTATTCGGATATCGTTAAGAAAGAATTCCCAGAGCATCTGAAAGAAGAACGGGTTGCCTGCATCCTCATGAACAACATTCAGCAGCTGCGAGTGCAGCTAGAGAAGATGTTCGAGGCGATGGGCGGCACCAAGCTGGAGCGCGACGCGGCCGACATCCTCCACGACTTGCAGCAGCGCCTCAACAGCACGCTGGACGAACTCGCATCCATGTTCGCTAATAGCCTGGAGTTCAGAATAACAGCGAGCGTGAAGGATCTAGGAGACCTGCTACAGAAGGTGGGCGGAGGCGGAGCGCCCGGCGTCCCACAGCACCCCGCGCACCAGGAGGCTGATGAGGTGCTCCGGCCCCTAATGGATATCTTGGACGGTTCTCTTACTATGTACGCGGAGTCTTGCGAGAAGACCGTCCTTAAGCGCCTTCTGAAGGAGCTGTGGAAGATTGTCATGAAGATATTAGAGAAAACCGTCGTACTCCCACCCATGACGGATAAAACGATGATGTTGAAGAACTTAACGAATAACGCCAAGAATTTAGCGGCGAACGCAAAGATAGAGGACATGACGCAGCTGTTCAAGAGCACCGTTGGCAAGCAGGACGTAAAGCACGCGCTCTCCGGCGTCATGGACATCTCTAAAGAACATCTGTCAGCCGAGAAGAGCTTAACGCCGAAGCAATGCGCGGTACTGGACGCGGCTCTGGACACCATCAAGCAGTACTTCCACGCCGGCGGCAACGGACTCAAGAAGACTTTCTTGGAAAAGAGTCCAGAGCTCCAGTCGCTGAGATACGCCCTCAGTCTGTACACGCAGACGACTGACACGTTGATCAGGACCTTCGTTACCAGCCAGCAGAATCAAGACGCCGCTGTTGCTGAGGAGGGAAGCGTCGGCGAAGTGTCCCTGCAAGTGGACCTCTTCACACATCCCGGGACGGGAGAACATAAAATCACTGTCAAAGTGGTTGCGGCAAATGATCTCAAGTGGGTGATCGCGAGTGGAATGTTCCGTCCGTTTATAGAGGTGAACCTGATAGGCCCTCACTTGGCCGACAAGAAGAGGAAGTTCGCCACCAAATCCAAGAGCAACAACTGGAGTCCCAAGTTCAATGAAACCTTCCATTTCATGGTGAGCGCCACTGAGCAGCTGGAGTTGTTCGAGCTGCACGTCTGCGTGCGGGACTACTGCTTCGCGCGCGAGGACCGGCTCGTCGGCGTCGCGGTCATGCGGCTCGCCGACATCGCGGAGCAGGGTTCGTGTGCCTGCTGGCTACCGCTCGGCACAAGGGTGAAGATGGACGAGACGGGCTGGACGATCCTAAGGATACTTTCTCAAAGACACAGCGACGAAGTGGCTAGGGAGTTCGTCAAATTAAAGTCGGAGATGAGAGCCGAGGCGCCGGCCGGAGGTCAAGGACCCTCGTGA
Protein
MSTLLANINAYYAHTTASSAVSASDRFAASNFGKEKFVKLLDQLHNSLRIDLSMYRNNFPASSAEKLMDLKSTVDLLTSITFFRMKVQELSSPPRASTVVKDCVKACLRSTYQFLFENCYELYNREFQVDPNEAKRDPADHGPQLDSVDFWHKLIALIVSVIEEDRNSYAPVLNQFPQELNIGQLSAATMWSLFAVDMKYALEEHEQHRLCKSSAYMNLHFKVKWLHTNYVKDVPPYCGAVPEYPAWFEPFVMQWLNENDDVSLEYLNGAFNRDKRDGFQKSSEHALFSNSVVDVFTQLTQCFDVVSKLECPDPEIWKRYMKRFAKTIVKVLVAYSDIVKKEFPEHLKEERVACILMNNIQQLRVQLEKMFEAMGGTKLERDAADILHDLQQRLNSTLDELASMFANSLEFRITASVKDLGDLLQKVGGGGAPGVPQHPAHQEADEVLRPLMDILDGSLTMYAESCEKTVLKRLLKELWKIVMKILEKTVVLPPMTDKTMMLKNLTNNAKNLAANAKIEDMTQLFKSTVGKQDVKHALSGVMDISKEHLSAEKSLTPKQCAVLDAALDTIKQYFHAGGNGLKKTFLEKSPELQSLRYALSLYTQTTDTLIRTFVTSQQNQDAAVAEEGSVGEVSLQVDLFTHPGTGEHKITVKVVAANDLKWVIASGMFRPFIEVNLIGPHLADKKRKFATKSKSNNWSPKFNETFHFMVSATEQLELFELHVCVRDYCFAREDRLVGVAVMRLADIAEQGSCACWLPLGTRVKMDETGWTILRILSQRHSDEVAREFVKLKSEMRAEAPAGGQGPS
Summary
Uniprot
A0A437BVV0
A0A194QI63
A0A194RIL4
A0A212EI03
A0A2A4JRG8
A0A2A4JQB9
+ More
A0A2H1VUI3 A0A1W4XHS5 A0A1Y1N5S8 A0A139W8Y1 A0A1W4XHR9 A0A1W4XHZ9 A0A1W4X7H7 A0A1W4XGT5 W8BQ47 A0A0K8USW3 A0A2J7R515 W8AZG6 A0A0K8WC69 A0A0K8U895 A0A3L8D8D4 A0A0Q9XQK7 A0A0M4F836 B4MF48 A0A0Q9WMM1 A0A0A1WWK8 A0A0Q9WW17 D0Z7E8 A0A0Q9WL03 A0A0Q9WL55 A0A0Q9WT43 A0A0Q9WUG5 A0A0J7L7R6 V9I9B5 A0A0A1XKS4 W8AS13 A0A146L4Q0 W8BEK5 A0A034VWT2 A0A1I8NRG3 E2B3A9 A0A0J9S108 A0A0J9USQ3 A0A0J9S110 Q8IM86 H9XVP2 A0A0R1E7I8 B4PW31 A0A0R1EC67 B4JZV1 L0MPR2 Q9V483 A0A0J9S2N7 Q8IM87 A0A1I8M4P3 A0A1W4V567 A0A1W4V4R1 X2J9E1 A0A1W4VIC1 A0A0J9S106 A0A0J9USP5 A0A0R1E6X5 A0A0J9S2P2 A0A0R1E6X9 A0A0J9S187 A0A0R1E742 A0A0R1E7J0 A0A1W4VHJ6 A0A0R1ECX6 A0A1W4VGY8 A0A0R1E8Q4 A0A0N8NZK0 A0A1W4V571 A0A3B0KQQ7 A0A0P8XYF4 A0A0P9A898 A0A0P8XI30 A0A0P9A891 A0A0P8XI65 A0A0P8XI37 A0A0N8NZJ9 A0A0P8XYG8 B3N047 B4HC83 E9IJC5 A0A1A9Y6D7 T1PL35 Q8T049 B4R2H5 A0A0P8XI97 A0A0P8XIA4 A0A0P5TJ17 A0A0P5ALP6 A0A0P5D6N2 A0A0P5TWX6 A0A0P6D701 A0A0P5CZR5 A0A0P5PTG6 A0A0P5JQH2 A0A0P5K2V8
A0A2H1VUI3 A0A1W4XHS5 A0A1Y1N5S8 A0A139W8Y1 A0A1W4XHR9 A0A1W4XHZ9 A0A1W4X7H7 A0A1W4XGT5 W8BQ47 A0A0K8USW3 A0A2J7R515 W8AZG6 A0A0K8WC69 A0A0K8U895 A0A3L8D8D4 A0A0Q9XQK7 A0A0M4F836 B4MF48 A0A0Q9WMM1 A0A0A1WWK8 A0A0Q9WW17 D0Z7E8 A0A0Q9WL03 A0A0Q9WL55 A0A0Q9WT43 A0A0Q9WUG5 A0A0J7L7R6 V9I9B5 A0A0A1XKS4 W8AS13 A0A146L4Q0 W8BEK5 A0A034VWT2 A0A1I8NRG3 E2B3A9 A0A0J9S108 A0A0J9USQ3 A0A0J9S110 Q8IM86 H9XVP2 A0A0R1E7I8 B4PW31 A0A0R1EC67 B4JZV1 L0MPR2 Q9V483 A0A0J9S2N7 Q8IM87 A0A1I8M4P3 A0A1W4V567 A0A1W4V4R1 X2J9E1 A0A1W4VIC1 A0A0J9S106 A0A0J9USP5 A0A0R1E6X5 A0A0J9S2P2 A0A0R1E6X9 A0A0J9S187 A0A0R1E742 A0A0R1E7J0 A0A1W4VHJ6 A0A0R1ECX6 A0A1W4VGY8 A0A0R1E8Q4 A0A0N8NZK0 A0A1W4V571 A0A3B0KQQ7 A0A0P8XYF4 A0A0P9A898 A0A0P8XI30 A0A0P9A891 A0A0P8XI65 A0A0P8XI37 A0A0N8NZJ9 A0A0P8XYG8 B3N047 B4HC83 E9IJC5 A0A1A9Y6D7 T1PL35 Q8T049 B4R2H5 A0A0P8XI97 A0A0P8XIA4 A0A0P5TJ17 A0A0P5ALP6 A0A0P5D6N2 A0A0P5TWX6 A0A0P6D701 A0A0P5CZR5 A0A0P5PTG6 A0A0P5JQH2 A0A0P5K2V8
Pubmed
EMBL
RSAL01000006
RVE54279.1
KQ459193
KPJ03141.1
KQ460325
KPJ15786.1
+ More
AGBW02014739 OWR41116.1 NWSH01000824 PCG74010.1 PCG74009.1 ODYU01004233 SOQ43894.1 GEZM01013030 JAV92808.1 KQ972879 KXZ75744.1 GAMC01014921 JAB91634.1 GDHF01022550 JAI29764.1 NEVH01007395 PNF35928.1 GAMC01014918 JAB91637.1 GDHF01003655 GDHF01002886 JAI48659.1 JAI49428.1 GDHF01029432 JAI22882.1 QOIP01000011 RLU16546.1 CH933813 KRG07246.1 CP012527 ALC48120.1 CH940665 EDW71149.2 KRF85446.1 GBXI01011397 JAD02895.1 KRF85439.1 KRF85441.1 KRF85442.1 KRF85444.1 KRF85450.1 CM000831 ACY70506.1 KRF85449.1 KRF85443.1 KRF85448.1 KRF85447.1 CH964272 KRF99807.1 LBMM01000328 KMR01208.1 JR036571 AEY57237.1 GBXI01003144 JAD11148.1 GAMC01014915 JAB91640.1 GDHC01015155 JAQ03474.1 GAMC01014919 GAMC01014917 JAB91636.1 GAKP01012652 JAC46300.1 GL445323 EFN89787.1 CM002916 KMZ07319.1 KMZ07326.1 KMZ07327.1 AE014135 AAN06593.1 AFH06787.1 CM000161 KRK04969.1 EDW99336.2 KRK04977.1 CH916380 EDV90967.1 AGB96565.1 AAF59405.4 KMZ07320.1 AAN06592.1 AFH06786.1 AHN58218.1 KMZ07322.1 KMZ07321.1 KMZ07329.1 KRK04976.1 KMZ07325.1 KRK04966.1 KMZ07323.1 KRK04973.1 KRK04965.1 KRK04968.1 KRK04972.1 KRK04970.1 KRK04974.1 CH902639 KPU74540.1 OUUW01000017 SPP88959.1 KPU74533.1 KPU74544.1 KPU74534.1 KPU74535.1 KPU74545.1 KPU74543.1 KPU74532.1 KPU74547.1 KPU74542.1 EDV33555.2 CH479282 EDW25237.1 GL763764 EFZ19343.1 KA648830 AFP63459.1 AY069559 AAL39704.1 CM000365 EDX15226.1 KPU74537.1 KPU74546.1 GDIQ01091070 JAL60656.1 GDIP01202107 JAJ21295.1 GDIP01164712 JAJ58690.1 GDIP01121467 JAL82247.1 GDIQ01081872 JAN12865.1 GDIP01168558 JAJ54844.1 GDIQ01123658 GDIQ01093418 GDIQ01091071 GDIQ01073127 JAL28068.1 GDIQ01195433 GDIQ01193535 JAK56292.1 GDIQ01195434 GDIQ01193534 JAK56291.1
AGBW02014739 OWR41116.1 NWSH01000824 PCG74010.1 PCG74009.1 ODYU01004233 SOQ43894.1 GEZM01013030 JAV92808.1 KQ972879 KXZ75744.1 GAMC01014921 JAB91634.1 GDHF01022550 JAI29764.1 NEVH01007395 PNF35928.1 GAMC01014918 JAB91637.1 GDHF01003655 GDHF01002886 JAI48659.1 JAI49428.1 GDHF01029432 JAI22882.1 QOIP01000011 RLU16546.1 CH933813 KRG07246.1 CP012527 ALC48120.1 CH940665 EDW71149.2 KRF85446.1 GBXI01011397 JAD02895.1 KRF85439.1 KRF85441.1 KRF85442.1 KRF85444.1 KRF85450.1 CM000831 ACY70506.1 KRF85449.1 KRF85443.1 KRF85448.1 KRF85447.1 CH964272 KRF99807.1 LBMM01000328 KMR01208.1 JR036571 AEY57237.1 GBXI01003144 JAD11148.1 GAMC01014915 JAB91640.1 GDHC01015155 JAQ03474.1 GAMC01014919 GAMC01014917 JAB91636.1 GAKP01012652 JAC46300.1 GL445323 EFN89787.1 CM002916 KMZ07319.1 KMZ07326.1 KMZ07327.1 AE014135 AAN06593.1 AFH06787.1 CM000161 KRK04969.1 EDW99336.2 KRK04977.1 CH916380 EDV90967.1 AGB96565.1 AAF59405.4 KMZ07320.1 AAN06592.1 AFH06786.1 AHN58218.1 KMZ07322.1 KMZ07321.1 KMZ07329.1 KRK04976.1 KMZ07325.1 KRK04966.1 KMZ07323.1 KRK04973.1 KRK04965.1 KRK04968.1 KRK04972.1 KRK04970.1 KRK04974.1 CH902639 KPU74540.1 OUUW01000017 SPP88959.1 KPU74533.1 KPU74544.1 KPU74534.1 KPU74535.1 KPU74545.1 KPU74543.1 KPU74532.1 KPU74547.1 KPU74542.1 EDV33555.2 CH479282 EDW25237.1 GL763764 EFZ19343.1 KA648830 AFP63459.1 AY069559 AAL39704.1 CM000365 EDX15226.1 KPU74537.1 KPU74546.1 GDIQ01091070 JAL60656.1 GDIP01202107 JAJ21295.1 GDIP01164712 JAJ58690.1 GDIP01121467 JAL82247.1 GDIQ01081872 JAN12865.1 GDIP01168558 JAJ54844.1 GDIQ01123658 GDIQ01093418 GDIQ01091071 GDIQ01073127 JAL28068.1 GDIQ01195433 GDIQ01193535 JAK56292.1 GDIQ01195434 GDIQ01193534 JAK56291.1
Proteomes
UP000283053
UP000053268
UP000053240
UP000007151
UP000218220
UP000192223
+ More
UP000007266 UP000235965 UP000279307 UP000009192 UP000092553 UP000008792 UP000007798 UP000036403 UP000095300 UP000008237 UP000000803 UP000002282 UP000001070 UP000095301 UP000192221 UP000007801 UP000268350 UP000008744 UP000092443 UP000000304
UP000007266 UP000235965 UP000279307 UP000009192 UP000092553 UP000008792 UP000007798 UP000036403 UP000095300 UP000008237 UP000000803 UP000002282 UP000001070 UP000095301 UP000192221 UP000007801 UP000268350 UP000008744 UP000092443 UP000000304
Interpro
Gene 3D
ProteinModelPortal
A0A437BVV0
A0A194QI63
A0A194RIL4
A0A212EI03
A0A2A4JRG8
A0A2A4JQB9
+ More
A0A2H1VUI3 A0A1W4XHS5 A0A1Y1N5S8 A0A139W8Y1 A0A1W4XHR9 A0A1W4XHZ9 A0A1W4X7H7 A0A1W4XGT5 W8BQ47 A0A0K8USW3 A0A2J7R515 W8AZG6 A0A0K8WC69 A0A0K8U895 A0A3L8D8D4 A0A0Q9XQK7 A0A0M4F836 B4MF48 A0A0Q9WMM1 A0A0A1WWK8 A0A0Q9WW17 D0Z7E8 A0A0Q9WL03 A0A0Q9WL55 A0A0Q9WT43 A0A0Q9WUG5 A0A0J7L7R6 V9I9B5 A0A0A1XKS4 W8AS13 A0A146L4Q0 W8BEK5 A0A034VWT2 A0A1I8NRG3 E2B3A9 A0A0J9S108 A0A0J9USQ3 A0A0J9S110 Q8IM86 H9XVP2 A0A0R1E7I8 B4PW31 A0A0R1EC67 B4JZV1 L0MPR2 Q9V483 A0A0J9S2N7 Q8IM87 A0A1I8M4P3 A0A1W4V567 A0A1W4V4R1 X2J9E1 A0A1W4VIC1 A0A0J9S106 A0A0J9USP5 A0A0R1E6X5 A0A0J9S2P2 A0A0R1E6X9 A0A0J9S187 A0A0R1E742 A0A0R1E7J0 A0A1W4VHJ6 A0A0R1ECX6 A0A1W4VGY8 A0A0R1E8Q4 A0A0N8NZK0 A0A1W4V571 A0A3B0KQQ7 A0A0P8XYF4 A0A0P9A898 A0A0P8XI30 A0A0P9A891 A0A0P8XI65 A0A0P8XI37 A0A0N8NZJ9 A0A0P8XYG8 B3N047 B4HC83 E9IJC5 A0A1A9Y6D7 T1PL35 Q8T049 B4R2H5 A0A0P8XI97 A0A0P8XIA4 A0A0P5TJ17 A0A0P5ALP6 A0A0P5D6N2 A0A0P5TWX6 A0A0P6D701 A0A0P5CZR5 A0A0P5PTG6 A0A0P5JQH2 A0A0P5K2V8
A0A2H1VUI3 A0A1W4XHS5 A0A1Y1N5S8 A0A139W8Y1 A0A1W4XHR9 A0A1W4XHZ9 A0A1W4X7H7 A0A1W4XGT5 W8BQ47 A0A0K8USW3 A0A2J7R515 W8AZG6 A0A0K8WC69 A0A0K8U895 A0A3L8D8D4 A0A0Q9XQK7 A0A0M4F836 B4MF48 A0A0Q9WMM1 A0A0A1WWK8 A0A0Q9WW17 D0Z7E8 A0A0Q9WL03 A0A0Q9WL55 A0A0Q9WT43 A0A0Q9WUG5 A0A0J7L7R6 V9I9B5 A0A0A1XKS4 W8AS13 A0A146L4Q0 W8BEK5 A0A034VWT2 A0A1I8NRG3 E2B3A9 A0A0J9S108 A0A0J9USQ3 A0A0J9S110 Q8IM86 H9XVP2 A0A0R1E7I8 B4PW31 A0A0R1EC67 B4JZV1 L0MPR2 Q9V483 A0A0J9S2N7 Q8IM87 A0A1I8M4P3 A0A1W4V567 A0A1W4V4R1 X2J9E1 A0A1W4VIC1 A0A0J9S106 A0A0J9USP5 A0A0R1E6X5 A0A0J9S2P2 A0A0R1E6X9 A0A0J9S187 A0A0R1E742 A0A0R1E7J0 A0A1W4VHJ6 A0A0R1ECX6 A0A1W4VGY8 A0A0R1E8Q4 A0A0N8NZK0 A0A1W4V571 A0A3B0KQQ7 A0A0P8XYF4 A0A0P9A898 A0A0P8XI30 A0A0P9A891 A0A0P8XI65 A0A0P8XI37 A0A0N8NZJ9 A0A0P8XYG8 B3N047 B4HC83 E9IJC5 A0A1A9Y6D7 T1PL35 Q8T049 B4R2H5 A0A0P8XI97 A0A0P8XIA4 A0A0P5TJ17 A0A0P5ALP6 A0A0P5D6N2 A0A0P5TWX6 A0A0P6D701 A0A0P5CZR5 A0A0P5PTG6 A0A0P5JQH2 A0A0P5K2V8
PDB
5UE8
E-value=0,
Score=1972
Ontologies
GO
GO:0019992
GO:0007268
GO:0035556
GO:0005509
GO:0005543
GO:0046872
GO:0099069
GO:0043195
GO:0045211
GO:0048786
GO:0016021
GO:0016079
GO:0008021
GO:0016082
GO:0007269
GO:0005516
GO:0005886
GO:0042734
GO:0017075
GO:0043005
GO:0005515
GO:0003676
GO:0007165
GO:0006418
GO:0004822
GO:0005737
GO:0006428
GO:0000049
GO:0006508
GO:0016286
PANTHER
Topology
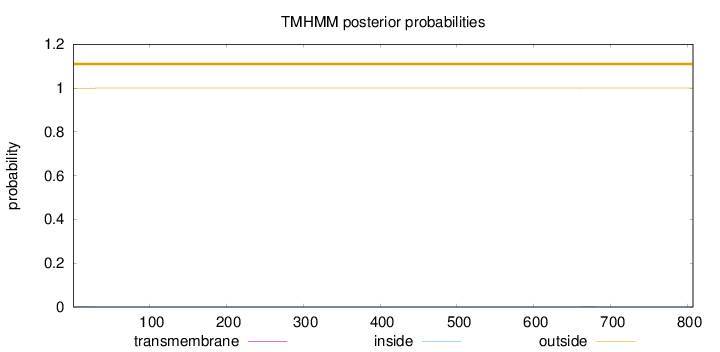
Length:
807
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02911
Exp number, first 60 AAs:
0.02495
Total prob of N-in:
0.00126
outside
1 - 807
Population Genetic Test Statistics
Pi
216.453183
Theta
184.603744
Tajima's D
0.986373
CLR
0.038583
CSRT
0.656767161641918
Interpretation
Uncertain
