Gene
KWMTBOMO12642
Pre Gene Modal
BGIBMGA001566
Annotation
PREDICTED:_protein_unc-13_homolog_A_isoform_X2_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.231
Sequence
CDS
ATGCCCGACATCAGGCCTCGAAGAAAATCGATCCCTCTTGTGTCAGAACTTACTATGGCTGCCACGAAAAGGAATGCTGGATTAACATCAGCGGTACATCGTGCAACACTTAATGATGAAGAGCTTAAAATGCACGTTTACAAGAAGACACTTCAAGCCCTCATCTACCCTATATCTTCAACAACCCCTCACAACTTTGTGCTGTGGACCGCCACTTCCCCTACTTATTGCTACGAGTGTGAAGGGCTTCTATGGGGTATCGCACGACAAGGGGTCAGGTGTACAGAGTGCGGGGTCAAATGCCATGAGAAATGCAAGGACTTGCTCAACGCTGATTGCCTTCAGATGTGTTTCGCAGGGGCTGCCGAGAAGTCTTCTAAGCACGGCGCGGAGGATAAAGCTAATTCGATTATAACAGCAATGAAGGAAAGAATGAAACAGCGAGAGCGAGACAAGCCCGAAATATTCGAACTCATCAGACGCGTGTTCAGCATCGAACAGGAGGGGCATGCAGCACACATGAGTACGGTGAAGCAATCAGTCCTCGATGGTACCTCGAAGTGGAGCGCGAAGATTGCCATAACAGTTATATGTGCTCAGGGTTTGATAGCGAAGGACAAGAGTGGGACCTCGGATCCTTACGTAACAGTTCAAGTAAGCAAAGTGAAGAAAAGGACTAGAACTATGCCTCAAGAACTGAACCCCGTTTGGAACGAGAAGTTTTACTTTGAGTGCCACAACTCCTCCGATCGAATCAAAGTGCGAGTGTGGGATGAAGACAATGATTTAAAATCGAAACTACGCCAAAAGTTAACCAGGGAATCTGACGACTTCTTAGGACAAACCATTATTGAGGTACGGACATTATCCGGTGAGATGGACGTTTGGTACAATCTGGAGAAGCGCACGGATAAATCAGCTGTATCCGGAGCTATAAGACTTCACATCAATGTAGAAATCAAGGGTGAAGAAAAGGTCGCTCCCTATCACGTACAACGTGAAACGTCAACAGGTTGCCTAAAATTGGCACCATGCCTTGGATTCACGCCAGTTTTGACCATAATCACAAATGTTGCATTCGACAAATTGAACTGCGAATGGTCTTCGACTAACTATCCGTGCAATTAA
Protein
MPDIRPRRKSIPLVSELTMAATKRNAGLTSAVHRATLNDEELKMHVYKKTLQALIYPISSTTPHNFVLWTATSPTYCYECEGLLWGIARQGVRCTECGVKCHEKCKDLLNADCLQMCFAGAAEKSSKHGAEDKANSIITAMKERMKQRERDKPEIFELIRRVFSIEQEGHAAHMSTVKQSVLDGTSKWSAKIAITVICAQGLIAKDKSGTSDPYVTVQVSKVKKRTRTMPQELNPVWNEKFYFECHNSSDRIKVRVWDEDNDLKSKLRQKLTRESDDFLGQTIIEVRTLSGEMDVWYNLEKRTDKSAVSGAIRLHINVEIKGEEKVAPYHVQRETSTGCLKLAPCLGFTPVLTIITNVAFDKLNCEWSSTNYPCN
Summary
Uniprot
A0A212EI05
A0A437BVV0
A0A194RER4
A0A194QI63
A0A2H1VI92
A0A182LD95
+ More
A0A182LYS0 A0A182YCL7 A0A340TB75 A0A182FJE6 A0A182JN52 A0A182PFU1 A0A182N016 A0A182VVN5 A0A182Q454 A0A182XEQ8 A0A2C9GPF9 A0A182V221 A0A182RZ41 B0WHF9 A0A182TIS8 A0A2C9GPF7 Q7QEM8 A0A084WPR8 A0A0K8USW3 A0A0K8WC69 A0A0K8TW57 A0A0K8U895 A0A0M4F836 D0Z7E8 A0A0Q9WL03 W8BQ47 A0A1W4VHJ6 A0A1W4V4R1 A0A3B0KQQ7 W8AZG6 Q8IM86 X2J9E1 A0A0J9USQ3 A0A0Q9WYC9 A0A0Q9XQK7 A0A0K8U894 A0A0J9S106 A0A0P9A891 A0A1I8M4P3 A0A1B0D855 A0A1I8NRG3 B4JZV1 A0A0Q9WL55 A0A0Q9WW17 B4MF48 B4PW31 A0A2A4JRG8 A0A1W4VGY8 A0A1W4VIC1 A0A1W4XGT5 A0A0J9S114 A0A1B0CAB3 H9XVP2 Q9V483 A0A0J9S110 A0A182H3A0 A0A0P8XYG8 B4HC83 A0A0P8XI30 A0A0P8XI65 A0A0P8XI37 A0A0Q9WUG5 A0A0J9S2N7 A0A0J9S2P2 A0A0R1E7M0 A0A0J9USP5 A0A0N8NZJ9 Q17IK9 A0A0R1EC67 T1PL35 A0A1W4X7H7 A0A0R1E6X9 A0A0R1E6X5 A0A0R1E8Q4 A0A0R1E742 A0A0R1E7J0 A0A0A1XKS4 A0A1Y1N5S8 O96960 O96959 V9I9T4 V9I9B5 A0A1J1J702 A0A158NEN0 A0A0P5ALZ6 A0A2A4JQB9 A0A0P5A4C5 A0A0P5ALP6 A0A0P6D701 A0A0P5TWX6 A0A0P5TJ17 A0A0P5PTG6 A0A0P5QAC9 A0A0P5TZW7
A0A182LYS0 A0A182YCL7 A0A340TB75 A0A182FJE6 A0A182JN52 A0A182PFU1 A0A182N016 A0A182VVN5 A0A182Q454 A0A182XEQ8 A0A2C9GPF9 A0A182V221 A0A182RZ41 B0WHF9 A0A182TIS8 A0A2C9GPF7 Q7QEM8 A0A084WPR8 A0A0K8USW3 A0A0K8WC69 A0A0K8TW57 A0A0K8U895 A0A0M4F836 D0Z7E8 A0A0Q9WL03 W8BQ47 A0A1W4VHJ6 A0A1W4V4R1 A0A3B0KQQ7 W8AZG6 Q8IM86 X2J9E1 A0A0J9USQ3 A0A0Q9WYC9 A0A0Q9XQK7 A0A0K8U894 A0A0J9S106 A0A0P9A891 A0A1I8M4P3 A0A1B0D855 A0A1I8NRG3 B4JZV1 A0A0Q9WL55 A0A0Q9WW17 B4MF48 B4PW31 A0A2A4JRG8 A0A1W4VGY8 A0A1W4VIC1 A0A1W4XGT5 A0A0J9S114 A0A1B0CAB3 H9XVP2 Q9V483 A0A0J9S110 A0A182H3A0 A0A0P8XYG8 B4HC83 A0A0P8XI30 A0A0P8XI65 A0A0P8XI37 A0A0Q9WUG5 A0A0J9S2N7 A0A0J9S2P2 A0A0R1E7M0 A0A0J9USP5 A0A0N8NZJ9 Q17IK9 A0A0R1EC67 T1PL35 A0A1W4X7H7 A0A0R1E6X9 A0A0R1E6X5 A0A0R1E8Q4 A0A0R1E742 A0A0R1E7J0 A0A0A1XKS4 A0A1Y1N5S8 O96960 O96959 V9I9T4 V9I9B5 A0A1J1J702 A0A158NEN0 A0A0P5ALZ6 A0A2A4JQB9 A0A0P5A4C5 A0A0P5ALP6 A0A0P6D701 A0A0P5TWX6 A0A0P5TJ17 A0A0P5PTG6 A0A0P5QAC9 A0A0P5TZW7
Pubmed
EMBL
AGBW02014738
OWR41118.1
RSAL01000006
RVE54279.1
KQ460325
KPJ15785.1
+ More
KQ459193 KPJ03141.1 ODYU01002710 SOQ40557.1 AXCM01003725 AXCN02001219 AXCN02001220 APCN01004044 APCN01004045 APCN01004046 DS231935 EDS27709.1 AAAB01008846 EAA06229.5 ATLV01025122 KE525369 KFB52212.1 GDHF01022550 JAI29764.1 GDHF01003655 GDHF01002886 JAI48659.1 JAI49428.1 GDHF01034024 GDHF01013408 JAI18290.1 JAI38906.1 GDHF01029432 JAI22882.1 CP012527 ALC48120.1 CM000831 ACY70506.1 CH940665 KRF85449.1 GAMC01014921 JAB91634.1 OUUW01000017 SPP88959.1 GAMC01014918 JAB91637.1 AE014135 AAN06593.1 AHN58218.1 CM002916 KMZ07326.1 KRF85440.1 CH933813 KRG07246.1 GDHF01029435 JAI22879.1 KMZ07322.1 CH902639 KPU74535.1 AJVK01027224 AJVK01027225 AJVK01027226 AJVK01027227 AJVK01027228 AJVK01027229 CH916380 EDV90967.1 KRF85443.1 KRF85448.1 KRF85439.1 KRF85441.1 KRF85442.1 KRF85444.1 KRF85450.1 EDW71149.2 CM000161 EDW99336.2 NWSH01000824 PCG74010.1 KMZ07324.1 AJWK01003606 AJWK01003607 AJWK01003608 AJWK01003609 AJWK01003610 AFH06787.1 AAF59405.4 KMZ07327.1 JXUM01106995 KQ565100 KXJ71334.1 KPU74542.1 CH479282 EDW25237.1 KPU74534.1 KPU74545.1 KPU74543.1 CH964272 KRF99807.1 KMZ07320.1 KMZ07325.1 KRK04975.1 KMZ07321.1 KMZ07329.1 KPU74532.1 KPU74547.1 CH477239 EAT46460.1 KRK04977.1 KA648830 AFP63459.1 KRK04966.1 KRK04976.1 KRK04974.1 KRK04973.1 KRK04965.1 KRK04968.1 KRK04972.1 GBXI01003144 JAD11148.1 GEZM01013030 JAV92808.1 Y17922 CAA76942.1 Y17921 CAA76941.1 JR036570 AEY57236.1 JR036571 AEY57237.1 CVRI01000074 CRL08168.1 ADTU01013540 ADTU01013541 GDIP01196912 JAJ26490.1 PCG74009.1 GDIP01204159 JAJ19243.1 GDIP01202107 JAJ21295.1 GDIQ01081872 JAN12865.1 GDIP01121467 JAL82247.1 GDIQ01091070 JAL60656.1 GDIQ01123658 GDIQ01093418 GDIQ01091071 GDIQ01073127 JAL28068.1 GDIQ01123660 JAL28066.1 GDIP01124115 JAL79599.1
KQ459193 KPJ03141.1 ODYU01002710 SOQ40557.1 AXCM01003725 AXCN02001219 AXCN02001220 APCN01004044 APCN01004045 APCN01004046 DS231935 EDS27709.1 AAAB01008846 EAA06229.5 ATLV01025122 KE525369 KFB52212.1 GDHF01022550 JAI29764.1 GDHF01003655 GDHF01002886 JAI48659.1 JAI49428.1 GDHF01034024 GDHF01013408 JAI18290.1 JAI38906.1 GDHF01029432 JAI22882.1 CP012527 ALC48120.1 CM000831 ACY70506.1 CH940665 KRF85449.1 GAMC01014921 JAB91634.1 OUUW01000017 SPP88959.1 GAMC01014918 JAB91637.1 AE014135 AAN06593.1 AHN58218.1 CM002916 KMZ07326.1 KRF85440.1 CH933813 KRG07246.1 GDHF01029435 JAI22879.1 KMZ07322.1 CH902639 KPU74535.1 AJVK01027224 AJVK01027225 AJVK01027226 AJVK01027227 AJVK01027228 AJVK01027229 CH916380 EDV90967.1 KRF85443.1 KRF85448.1 KRF85439.1 KRF85441.1 KRF85442.1 KRF85444.1 KRF85450.1 EDW71149.2 CM000161 EDW99336.2 NWSH01000824 PCG74010.1 KMZ07324.1 AJWK01003606 AJWK01003607 AJWK01003608 AJWK01003609 AJWK01003610 AFH06787.1 AAF59405.4 KMZ07327.1 JXUM01106995 KQ565100 KXJ71334.1 KPU74542.1 CH479282 EDW25237.1 KPU74534.1 KPU74545.1 KPU74543.1 CH964272 KRF99807.1 KMZ07320.1 KMZ07325.1 KRK04975.1 KMZ07321.1 KMZ07329.1 KPU74532.1 KPU74547.1 CH477239 EAT46460.1 KRK04977.1 KA648830 AFP63459.1 KRK04966.1 KRK04976.1 KRK04974.1 KRK04973.1 KRK04965.1 KRK04968.1 KRK04972.1 GBXI01003144 JAD11148.1 GEZM01013030 JAV92808.1 Y17922 CAA76942.1 Y17921 CAA76941.1 JR036570 AEY57236.1 JR036571 AEY57237.1 CVRI01000074 CRL08168.1 ADTU01013540 ADTU01013541 GDIP01196912 JAJ26490.1 PCG74009.1 GDIP01204159 JAJ19243.1 GDIP01202107 JAJ21295.1 GDIQ01081872 JAN12865.1 GDIP01121467 JAL82247.1 GDIQ01091070 JAL60656.1 GDIQ01123658 GDIQ01093418 GDIQ01091071 GDIQ01073127 JAL28068.1 GDIQ01123660 JAL28066.1 GDIP01124115 JAL79599.1
Proteomes
UP000007151
UP000283053
UP000053240
UP000053268
UP000075882
UP000075883
+ More
UP000076408 UP000075903 UP000069272 UP000075880 UP000075885 UP000075884 UP000075920 UP000075886 UP000076407 UP000075840 UP000075900 UP000002320 UP000075902 UP000007062 UP000030765 UP000092553 UP000008792 UP000192221 UP000268350 UP000000803 UP000009192 UP000007801 UP000095301 UP000092462 UP000095300 UP000001070 UP000002282 UP000218220 UP000192223 UP000092461 UP000069940 UP000249989 UP000008744 UP000007798 UP000008820 UP000183832 UP000005205
UP000076408 UP000075903 UP000069272 UP000075880 UP000075885 UP000075884 UP000075920 UP000075886 UP000076407 UP000075840 UP000075900 UP000002320 UP000075902 UP000007062 UP000030765 UP000092553 UP000008792 UP000192221 UP000268350 UP000000803 UP000009192 UP000007801 UP000095301 UP000092462 UP000095300 UP000001070 UP000002282 UP000218220 UP000192223 UP000092461 UP000069940 UP000249989 UP000008744 UP000007798 UP000008820 UP000183832 UP000005205
Pfam
Interpro
IPR027080
Unc-13
+ More
IPR035892 C2_domain_sf
IPR037302 Unc-13_C2B
IPR002219 PE/DAG-bd
IPR000008 C2_dom
IPR014772 Munc13_dom-2
IPR014770 Munc13_1
IPR019558 Munc13_subgr_dom-2
IPR010439 CAPS_dom
IPR024733 NAGLU_tim-barrel
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR036116 FN3_sf
IPR021012 Dscam_C
IPR013098 Ig_I-set
IPR013151 Immunoglobulin
IPR013783 Ig-like_fold
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR003961 FN3_dom
IPR035892 C2_domain_sf
IPR037302 Unc-13_C2B
IPR002219 PE/DAG-bd
IPR000008 C2_dom
IPR014772 Munc13_dom-2
IPR014770 Munc13_1
IPR019558 Munc13_subgr_dom-2
IPR010439 CAPS_dom
IPR024733 NAGLU_tim-barrel
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR036116 FN3_sf
IPR021012 Dscam_C
IPR013098 Ig_I-set
IPR013151 Immunoglobulin
IPR013783 Ig-like_fold
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR003961 FN3_dom
Gene 3D
ProteinModelPortal
A0A212EI05
A0A437BVV0
A0A194RER4
A0A194QI63
A0A2H1VI92
A0A182LD95
+ More
A0A182LYS0 A0A182YCL7 A0A340TB75 A0A182FJE6 A0A182JN52 A0A182PFU1 A0A182N016 A0A182VVN5 A0A182Q454 A0A182XEQ8 A0A2C9GPF9 A0A182V221 A0A182RZ41 B0WHF9 A0A182TIS8 A0A2C9GPF7 Q7QEM8 A0A084WPR8 A0A0K8USW3 A0A0K8WC69 A0A0K8TW57 A0A0K8U895 A0A0M4F836 D0Z7E8 A0A0Q9WL03 W8BQ47 A0A1W4VHJ6 A0A1W4V4R1 A0A3B0KQQ7 W8AZG6 Q8IM86 X2J9E1 A0A0J9USQ3 A0A0Q9WYC9 A0A0Q9XQK7 A0A0K8U894 A0A0J9S106 A0A0P9A891 A0A1I8M4P3 A0A1B0D855 A0A1I8NRG3 B4JZV1 A0A0Q9WL55 A0A0Q9WW17 B4MF48 B4PW31 A0A2A4JRG8 A0A1W4VGY8 A0A1W4VIC1 A0A1W4XGT5 A0A0J9S114 A0A1B0CAB3 H9XVP2 Q9V483 A0A0J9S110 A0A182H3A0 A0A0P8XYG8 B4HC83 A0A0P8XI30 A0A0P8XI65 A0A0P8XI37 A0A0Q9WUG5 A0A0J9S2N7 A0A0J9S2P2 A0A0R1E7M0 A0A0J9USP5 A0A0N8NZJ9 Q17IK9 A0A0R1EC67 T1PL35 A0A1W4X7H7 A0A0R1E6X9 A0A0R1E6X5 A0A0R1E8Q4 A0A0R1E742 A0A0R1E7J0 A0A0A1XKS4 A0A1Y1N5S8 O96960 O96959 V9I9T4 V9I9B5 A0A1J1J702 A0A158NEN0 A0A0P5ALZ6 A0A2A4JQB9 A0A0P5A4C5 A0A0P5ALP6 A0A0P6D701 A0A0P5TWX6 A0A0P5TJ17 A0A0P5PTG6 A0A0P5QAC9 A0A0P5TZW7
A0A182LYS0 A0A182YCL7 A0A340TB75 A0A182FJE6 A0A182JN52 A0A182PFU1 A0A182N016 A0A182VVN5 A0A182Q454 A0A182XEQ8 A0A2C9GPF9 A0A182V221 A0A182RZ41 B0WHF9 A0A182TIS8 A0A2C9GPF7 Q7QEM8 A0A084WPR8 A0A0K8USW3 A0A0K8WC69 A0A0K8TW57 A0A0K8U895 A0A0M4F836 D0Z7E8 A0A0Q9WL03 W8BQ47 A0A1W4VHJ6 A0A1W4V4R1 A0A3B0KQQ7 W8AZG6 Q8IM86 X2J9E1 A0A0J9USQ3 A0A0Q9WYC9 A0A0Q9XQK7 A0A0K8U894 A0A0J9S106 A0A0P9A891 A0A1I8M4P3 A0A1B0D855 A0A1I8NRG3 B4JZV1 A0A0Q9WL55 A0A0Q9WW17 B4MF48 B4PW31 A0A2A4JRG8 A0A1W4VGY8 A0A1W4VIC1 A0A1W4XGT5 A0A0J9S114 A0A1B0CAB3 H9XVP2 Q9V483 A0A0J9S110 A0A182H3A0 A0A0P8XYG8 B4HC83 A0A0P8XI30 A0A0P8XI65 A0A0P8XI37 A0A0Q9WUG5 A0A0J9S2N7 A0A0J9S2P2 A0A0R1E7M0 A0A0J9USP5 A0A0N8NZJ9 Q17IK9 A0A0R1EC67 T1PL35 A0A1W4X7H7 A0A0R1E6X9 A0A0R1E6X5 A0A0R1E8Q4 A0A0R1E742 A0A0R1E7J0 A0A0A1XKS4 A0A1Y1N5S8 O96960 O96959 V9I9T4 V9I9B5 A0A1J1J702 A0A158NEN0 A0A0P5ALZ6 A0A2A4JQB9 A0A0P5A4C5 A0A0P5ALP6 A0A0P6D701 A0A0P5TWX6 A0A0P5TJ17 A0A0P5PTG6 A0A0P5QAC9 A0A0P5TZW7
PDB
5UE8
E-value=3.07145e-144,
Score=1312
Ontologies
GO
GO:0019992
GO:0007268
GO:0035556
GO:0005543
GO:0005509
GO:0005886
GO:0048786
GO:0042734
GO:0017075
GO:0031594
GO:0035249
GO:0061789
GO:0043195
GO:0016081
GO:0099525
GO:0030672
GO:0016188
GO:0016082
GO:0098831
GO:0007528
GO:0005516
GO:0045211
GO:0016079
GO:0008021
GO:0007269
GO:0043005
GO:0099069
GO:0016021
GO:0005515
GO:0003676
GO:0007165
GO:0006418
GO:0004822
GO:0005737
GO:0006428
GO:0000049
GO:0006508
GO:0016286
PANTHER
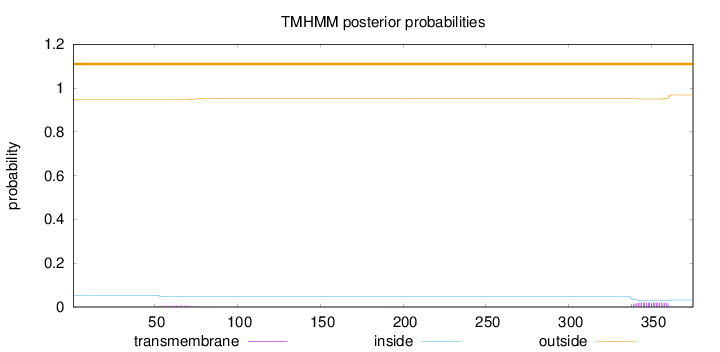
Topology
Length:
375
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.56599
Exp number, first 60 AAs:
0.04425
Total prob of N-in:
0.05361
outside
1 - 375
Population Genetic Test Statistics
Pi
186.656533
Theta
187.524746
Tajima's D
0.433727
CLR
0.073041
CSRT
0.502424878756062
Interpretation
Uncertain
