Gene
KWMTBOMO12627
Annotation
PREDICTED:_uncharacterized_protein_LOC106129721_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.276
Sequence
CDS
ATGTCTGCGACCTGGATAAGAATCACTCCGGGGTTAAGTAAAGGAATTCATGTCATCAATCTACTCGAGTTGTCACGTTTTGAGCAATTTTTAAATCGCATCCTAGTTAAACTAAAAATGAATAATAATGAAGTATTTTCACAAGAAGAACAAGAAAAGCTCACAAAATTGTTTAAAGTCGACGAAGAAAGCTTAGTAATGAGTATTAAAACATTGAAATATTTATTAAAAAAAATGTTGAAATTCATTTTTGTACCACTAACATTAAGAACTGATTTAAAAAGTCTTGGATTGTGTGATGAAAAGGCAGATGTTATTGTTAAAGTATGGTCTTCCGAAACGAGGCCAATACTTGATCAATTCAGTAAAGAAACTTCTGGTATGTCACAGGATGACTTAAATTTCACATGGAAATTAAATGCAGAGTTGAGTTCTGATTATTGCAAAAAAACAAAAATCCCTAAAGCATATTTGTCTCTAACCAATCATAAAGGCCATACTGAAATAGAATTGACACATTCAGAGTTATATTCCATGTTTATTAATTTTGAACAGATACAAAATGAATTGGATAATTTATAA
Protein
MSATWIRITPGLSKGIHVINLLELSRFEQFLNRILVKLKMNNNEVFSQEEQEKLTKLFKVDEESLVMSIKTLKYLLKKMLKFIFVPLTLRTDLKSLGLCDEKADVIVKVWSSETRPILDQFSKETSGMSQDDLNFTWKLNAELSSDYCKKTKIPKAYLSLTNHKGHTEIELTHSELYSMFINFEQIQNELDNL
Summary
Uniprot
A0A2H1W1Z1
A0A2A4IZL4
S4PWZ4
A0A212EST9
A0A194RE80
A0A3S2NLX5
+ More
A0A0N1INC2 A0A1S3JWK8 A0A293M367 A0A2R5LD30 A0A2Z4HQN6 A0A0K8TR52 W4YY82 A0A2J7QK51 C3ZA46 A0A0L8HS12 L7LVT7 A0A131YT28 T1GT82 F6UBQ5 A0A3Q0JUR5 A0A131XJ09 A0A210QYZ1 A0A034WJ23 A0A067QU80 V4ABV9 A0A224YD76 A0A0K8WEZ4 W8B4Y6 A0A1E1XJL8 H0VW16 A7SUC8 K9IHM3 A0A310SRY5 H2Z3L1 A0A1I8PD31 B3RRB6 A0A183U129 R7U9R6 A0A0L0BTU2 A0A1L8EB57 A0A341B8E5 A0A2Y9PMD1 A0A3M6TC65 A0A1A9WVV8 A0A2U4BTF0 A0A1A9X7W4 A0A1B0ALR0 A0A0B2UY21 A0A1A9UUA8 A0A023FA02 A0A2K6EZ29 A0A0L7R7R6 A0A3Q2PX04 A0A340WUL0 A0A369SE27 A0A088AJ65 A0A087X9G4 A0A3B3TZQ2 H1A4L1 A0A026W188 L7LUE8 A0A2Y9I0Z5 A0A2Y9SJJ9 A0A3Q2XGV8 A0A147A1Q8 A0A3B3YDL5 F6U5L1 E2B918 A0A287AYH8 A0A1I8MTN8 T1PIU4 A0A087UWK1 F1LCY8 A0A0M3HU34 A0A3Q7ND85 A0A2U3W3N7 A0A1X7U0Y5 M3XNZ6 A0A3Q7WPP5 A0A0P4VQR4 K4FV47 A0A1W5ADP5 A0A2R9ALT5 A0A0S7LSA3 A0A3B5KKL9 A0A1S2ZNE0 A0A3M7REX0 A0A3B4BV51 A0A2K5MCK8 U6CTQ6 A0A0P6K171 A0A2K5XFN0 A0A3Q2EEG7 A0A3B3DU96 A0A2T7NF21 A0A2Y9DMP2 A0A1B6M6W4 F6W770
A0A0N1INC2 A0A1S3JWK8 A0A293M367 A0A2R5LD30 A0A2Z4HQN6 A0A0K8TR52 W4YY82 A0A2J7QK51 C3ZA46 A0A0L8HS12 L7LVT7 A0A131YT28 T1GT82 F6UBQ5 A0A3Q0JUR5 A0A131XJ09 A0A210QYZ1 A0A034WJ23 A0A067QU80 V4ABV9 A0A224YD76 A0A0K8WEZ4 W8B4Y6 A0A1E1XJL8 H0VW16 A7SUC8 K9IHM3 A0A310SRY5 H2Z3L1 A0A1I8PD31 B3RRB6 A0A183U129 R7U9R6 A0A0L0BTU2 A0A1L8EB57 A0A341B8E5 A0A2Y9PMD1 A0A3M6TC65 A0A1A9WVV8 A0A2U4BTF0 A0A1A9X7W4 A0A1B0ALR0 A0A0B2UY21 A0A1A9UUA8 A0A023FA02 A0A2K6EZ29 A0A0L7R7R6 A0A3Q2PX04 A0A340WUL0 A0A369SE27 A0A088AJ65 A0A087X9G4 A0A3B3TZQ2 H1A4L1 A0A026W188 L7LUE8 A0A2Y9I0Z5 A0A2Y9SJJ9 A0A3Q2XGV8 A0A147A1Q8 A0A3B3YDL5 F6U5L1 E2B918 A0A287AYH8 A0A1I8MTN8 T1PIU4 A0A087UWK1 F1LCY8 A0A0M3HU34 A0A3Q7ND85 A0A2U3W3N7 A0A1X7U0Y5 M3XNZ6 A0A3Q7WPP5 A0A0P4VQR4 K4FV47 A0A1W5ADP5 A0A2R9ALT5 A0A0S7LSA3 A0A3B5KKL9 A0A1S2ZNE0 A0A3M7REX0 A0A3B4BV51 A0A2K5MCK8 U6CTQ6 A0A0P6K171 A0A2K5XFN0 A0A3Q2EEG7 A0A3B3DU96 A0A2T7NF21 A0A2Y9DMP2 A0A1B6M6W4 F6W770
Pubmed
23622113
22118469
26354079
29551666
26369729
18563158
+ More
25576852 26830274 12481130 15114417 28049606 28812685 25348373 24845553 23254933 28797301 24495485 29209593 21993624 17615350 18719581 26108605 30382153 25474469 30042472 20360741 24508170 30249741 19892987 20798317 30723633 25315136 21685128 27129103 23056606 22722832 21551351 30375419 29451363 25243066
25576852 26830274 12481130 15114417 28049606 28812685 25348373 24845553 23254933 28797301 24495485 29209593 21993624 17615350 18719581 26108605 30382153 25474469 30042472 20360741 24508170 30249741 19892987 20798317 30723633 25315136 21685128 27129103 23056606 22722832 21551351 30375419 29451363 25243066
EMBL
ODYU01005827
SOQ47101.1
NWSH01004601
PCG64868.1
GAIX01005933
JAA86627.1
+ More
AGBW02012716 OWR44566.1 KQ460325 KPJ15774.1 RSAL01000006 RVE54290.1 KQ458751 KPJ05306.1 GFWV01010684 GFWV01011917 MAA35413.1 GGLE01003308 MBY07434.1 MF579244 AWW17682.1 GDAI01001002 JAI16601.1 AAGJ04018162 NEVH01013278 PNF28964.1 GG666601 EEN50634.1 KQ417430 KOF92001.1 GACK01008853 JAA56181.1 GEDV01006184 JAP82373.1 CAQQ02200218 EAAA01000453 GEFH01002915 JAP65666.1 NEDP02001165 OWF53988.1 GAKP01005189 JAC53763.1 KK852936 KDR13576.1 KB202050 ESO92570.1 GFPF01000778 MAA11924.1 GDHF01002596 JAI49718.1 GAMC01021701 JAB84854.1 GFAA01004118 JAT99316.1 AAKN02023054 AAKN02023055 AAKN02023056 AAKN02023057 AAKN02023058 AAKN02023059 AAKN02023060 AAKN02023061 AAKN02023062 DS469811 EDO32681.1 GABZ01007988 JAA45537.1 KQ760712 OAD59305.1 DS985243 EDV26314.1 UYWY01002070 VDM27642.1 AMQN01008672 KB303698 ELU02871.1 JRES01001349 KNC23467.1 GFDG01002905 JAV15894.1 RCHS01003909 RMX38960.1 JXJN01000077 JPKZ01002973 KHN74104.1 GBBI01000396 JAC18316.1 KQ414637 KOC66927.1 NOWV01000018 RDD45175.1 AYCK01006961 ABQF01050470 ABQF01050471 ABQF01050472 ABQF01050473 ABQF01050474 KK107499 QOIP01000003 EZA49778.1 RLU25033.1 GACK01010516 JAA54518.1 GCES01014156 JAR72167.1 GL446425 EFN87784.1 AEMK02000012 DQIR01109189 DQIR01204367 HDA64665.1 HDB59844.1 KA648727 AFP63356.1 KK122009 KFM81740.1 JI178209 ADY47992.1 AEYP01047981 AEYP01047982 AEYP01047983 AEYP01047984 AEYP01047985 AEYP01047986 AEYP01047987 GDKW01001234 JAI55361.1 JX053371 AFK11599.1 AJFE02093652 GBYX01152156 JAO92298.1 REGN01003546 RNA22067.1 HAAF01001873 CCP73699.1 GEBF01007681 JAN95951.1 PZQS01000013 PVD19722.1 GEBQ01008336 JAT31641.1 GAMT01001122 GAMS01007822 GAMQ01005142 GAMP01007396 JAB10739.1 JAB15314.1 JAB36709.1 JAB45359.1
AGBW02012716 OWR44566.1 KQ460325 KPJ15774.1 RSAL01000006 RVE54290.1 KQ458751 KPJ05306.1 GFWV01010684 GFWV01011917 MAA35413.1 GGLE01003308 MBY07434.1 MF579244 AWW17682.1 GDAI01001002 JAI16601.1 AAGJ04018162 NEVH01013278 PNF28964.1 GG666601 EEN50634.1 KQ417430 KOF92001.1 GACK01008853 JAA56181.1 GEDV01006184 JAP82373.1 CAQQ02200218 EAAA01000453 GEFH01002915 JAP65666.1 NEDP02001165 OWF53988.1 GAKP01005189 JAC53763.1 KK852936 KDR13576.1 KB202050 ESO92570.1 GFPF01000778 MAA11924.1 GDHF01002596 JAI49718.1 GAMC01021701 JAB84854.1 GFAA01004118 JAT99316.1 AAKN02023054 AAKN02023055 AAKN02023056 AAKN02023057 AAKN02023058 AAKN02023059 AAKN02023060 AAKN02023061 AAKN02023062 DS469811 EDO32681.1 GABZ01007988 JAA45537.1 KQ760712 OAD59305.1 DS985243 EDV26314.1 UYWY01002070 VDM27642.1 AMQN01008672 KB303698 ELU02871.1 JRES01001349 KNC23467.1 GFDG01002905 JAV15894.1 RCHS01003909 RMX38960.1 JXJN01000077 JPKZ01002973 KHN74104.1 GBBI01000396 JAC18316.1 KQ414637 KOC66927.1 NOWV01000018 RDD45175.1 AYCK01006961 ABQF01050470 ABQF01050471 ABQF01050472 ABQF01050473 ABQF01050474 KK107499 QOIP01000003 EZA49778.1 RLU25033.1 GACK01010516 JAA54518.1 GCES01014156 JAR72167.1 GL446425 EFN87784.1 AEMK02000012 DQIR01109189 DQIR01204367 HDA64665.1 HDB59844.1 KA648727 AFP63356.1 KK122009 KFM81740.1 JI178209 ADY47992.1 AEYP01047981 AEYP01047982 AEYP01047983 AEYP01047984 AEYP01047985 AEYP01047986 AEYP01047987 GDKW01001234 JAI55361.1 JX053371 AFK11599.1 AJFE02093652 GBYX01152156 JAO92298.1 REGN01003546 RNA22067.1 HAAF01001873 CCP73699.1 GEBF01007681 JAN95951.1 PZQS01000013 PVD19722.1 GEBQ01008336 JAT31641.1 GAMT01001122 GAMS01007822 GAMQ01005142 GAMP01007396 JAB10739.1 JAB15314.1 JAB36709.1 JAB45359.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000283053
UP000053268
UP000085678
+ More
UP000007110 UP000235965 UP000001554 UP000053454 UP000015102 UP000008144 UP000242188 UP000027135 UP000030746 UP000005447 UP000001593 UP000007875 UP000095300 UP000009022 UP000050794 UP000267007 UP000014760 UP000037069 UP000252040 UP000248483 UP000275408 UP000091820 UP000245320 UP000092443 UP000092460 UP000031036 UP000078200 UP000233160 UP000053825 UP000265000 UP000265300 UP000253843 UP000005203 UP000028760 UP000261500 UP000007754 UP000053097 UP000279307 UP000248481 UP000248484 UP000264820 UP000261480 UP000002281 UP000008237 UP000008227 UP000095301 UP000054359 UP000036681 UP000286641 UP000245340 UP000007879 UP000000715 UP000286642 UP000192224 UP000240080 UP000005226 UP000079721 UP000276133 UP000261440 UP000233060 UP000233140 UP000265020 UP000261560 UP000245119 UP000248480
UP000007110 UP000235965 UP000001554 UP000053454 UP000015102 UP000008144 UP000242188 UP000027135 UP000030746 UP000005447 UP000001593 UP000007875 UP000095300 UP000009022 UP000050794 UP000267007 UP000014760 UP000037069 UP000252040 UP000248483 UP000275408 UP000091820 UP000245320 UP000092443 UP000092460 UP000031036 UP000078200 UP000233160 UP000053825 UP000265000 UP000265300 UP000253843 UP000005203 UP000028760 UP000261500 UP000007754 UP000053097 UP000279307 UP000248481 UP000248484 UP000264820 UP000261480 UP000002281 UP000008237 UP000008227 UP000095301 UP000054359 UP000036681 UP000286641 UP000245340 UP000007879 UP000000715 UP000286642 UP000192224 UP000240080 UP000005226 UP000079721 UP000276133 UP000261440 UP000233060 UP000233140 UP000265020 UP000261560 UP000245119 UP000248480
Pfam
PF07258 COMM_domain
CDD
ProteinModelPortal
A0A2H1W1Z1
A0A2A4IZL4
S4PWZ4
A0A212EST9
A0A194RE80
A0A3S2NLX5
+ More
A0A0N1INC2 A0A1S3JWK8 A0A293M367 A0A2R5LD30 A0A2Z4HQN6 A0A0K8TR52 W4YY82 A0A2J7QK51 C3ZA46 A0A0L8HS12 L7LVT7 A0A131YT28 T1GT82 F6UBQ5 A0A3Q0JUR5 A0A131XJ09 A0A210QYZ1 A0A034WJ23 A0A067QU80 V4ABV9 A0A224YD76 A0A0K8WEZ4 W8B4Y6 A0A1E1XJL8 H0VW16 A7SUC8 K9IHM3 A0A310SRY5 H2Z3L1 A0A1I8PD31 B3RRB6 A0A183U129 R7U9R6 A0A0L0BTU2 A0A1L8EB57 A0A341B8E5 A0A2Y9PMD1 A0A3M6TC65 A0A1A9WVV8 A0A2U4BTF0 A0A1A9X7W4 A0A1B0ALR0 A0A0B2UY21 A0A1A9UUA8 A0A023FA02 A0A2K6EZ29 A0A0L7R7R6 A0A3Q2PX04 A0A340WUL0 A0A369SE27 A0A088AJ65 A0A087X9G4 A0A3B3TZQ2 H1A4L1 A0A026W188 L7LUE8 A0A2Y9I0Z5 A0A2Y9SJJ9 A0A3Q2XGV8 A0A147A1Q8 A0A3B3YDL5 F6U5L1 E2B918 A0A287AYH8 A0A1I8MTN8 T1PIU4 A0A087UWK1 F1LCY8 A0A0M3HU34 A0A3Q7ND85 A0A2U3W3N7 A0A1X7U0Y5 M3XNZ6 A0A3Q7WPP5 A0A0P4VQR4 K4FV47 A0A1W5ADP5 A0A2R9ALT5 A0A0S7LSA3 A0A3B5KKL9 A0A1S2ZNE0 A0A3M7REX0 A0A3B4BV51 A0A2K5MCK8 U6CTQ6 A0A0P6K171 A0A2K5XFN0 A0A3Q2EEG7 A0A3B3DU96 A0A2T7NF21 A0A2Y9DMP2 A0A1B6M6W4 F6W770
A0A0N1INC2 A0A1S3JWK8 A0A293M367 A0A2R5LD30 A0A2Z4HQN6 A0A0K8TR52 W4YY82 A0A2J7QK51 C3ZA46 A0A0L8HS12 L7LVT7 A0A131YT28 T1GT82 F6UBQ5 A0A3Q0JUR5 A0A131XJ09 A0A210QYZ1 A0A034WJ23 A0A067QU80 V4ABV9 A0A224YD76 A0A0K8WEZ4 W8B4Y6 A0A1E1XJL8 H0VW16 A7SUC8 K9IHM3 A0A310SRY5 H2Z3L1 A0A1I8PD31 B3RRB6 A0A183U129 R7U9R6 A0A0L0BTU2 A0A1L8EB57 A0A341B8E5 A0A2Y9PMD1 A0A3M6TC65 A0A1A9WVV8 A0A2U4BTF0 A0A1A9X7W4 A0A1B0ALR0 A0A0B2UY21 A0A1A9UUA8 A0A023FA02 A0A2K6EZ29 A0A0L7R7R6 A0A3Q2PX04 A0A340WUL0 A0A369SE27 A0A088AJ65 A0A087X9G4 A0A3B3TZQ2 H1A4L1 A0A026W188 L7LUE8 A0A2Y9I0Z5 A0A2Y9SJJ9 A0A3Q2XGV8 A0A147A1Q8 A0A3B3YDL5 F6U5L1 E2B918 A0A287AYH8 A0A1I8MTN8 T1PIU4 A0A087UWK1 F1LCY8 A0A0M3HU34 A0A3Q7ND85 A0A2U3W3N7 A0A1X7U0Y5 M3XNZ6 A0A3Q7WPP5 A0A0P4VQR4 K4FV47 A0A1W5ADP5 A0A2R9ALT5 A0A0S7LSA3 A0A3B5KKL9 A0A1S2ZNE0 A0A3M7REX0 A0A3B4BV51 A0A2K5MCK8 U6CTQ6 A0A0P6K171 A0A2K5XFN0 A0A3Q2EEG7 A0A3B3DU96 A0A2T7NF21 A0A2Y9DMP2 A0A1B6M6W4 F6W770
Ontologies
GO
PANTHER
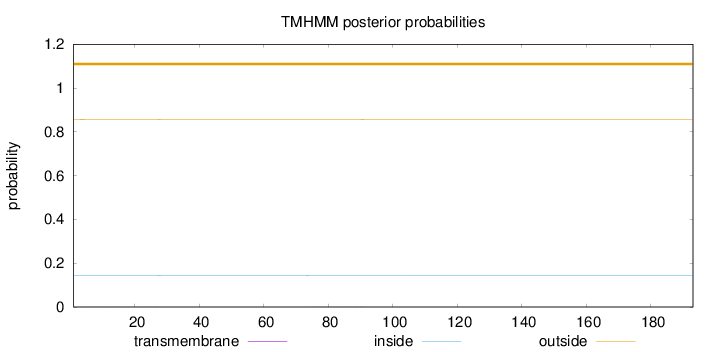
Topology
Length:
193
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02999
Exp number, first 60 AAs:
0.02461
Total prob of N-in:
0.14304
outside
1 - 193
Population Genetic Test Statistics
Pi
41.031188
Theta
69.556613
Tajima's D
-1.659985
CLR
101.954942
CSRT
0.0444477776111194
Interpretation
Uncertain
