Gene
KWMTBOMO12626 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001417
Annotation
PREDICTED:_protein_ariadne-1_[Papilio_polytes]
Full name
RBR-type E3 ubiquitin transferase
Location in the cell
Nuclear Reliability : 3.697
Sequence
CDS
ATGGATTCGGAAGATGACACGAAAGACGATGTGGACTCGGGTAATGAGTCTAGTGGAGACGATGTCGACTTTGCAATGGACGTTGAAACTCATAGTACAAGGGAAAGACAAACTGAGCTTGAGGAGTATCCTTACGAGGTTTTGTCTACAGAGGAGATTGTCCAACATATGGTTGACTGCATTAAAGAAGTGAACACTGTCGTCGAGATTCCAGCAACAACAACAAGGATCTTACTGAATCATTTCAAATGGGACAAAGAGAAGCTTATGGAACGTTTTTATGATGGAGACCAGGATCAGTTGTTCTCTGAAGCTAGAGTTATCAATCCATTCAGGAAACCTGTCATACAAAGGCCAAAGCTTCCCAGGAGGATATCAACGTCTGGTATGGAAGAATGTGAAATATGTTTCTCAGTTTTACCAACCTCTATTATGACTGGGTTGGAATGTGGTCACAGATTTTGCACGCAATGCTGGTGTGAATATTTAACTACGAAAATAATGGAAGAAGGCTTAGGTCAAACAATAGCTTGTGCCGCTCACGCCTGCGACATACTTGTTGACGATGCGACCGTGATGCGGCTAGTACGAGACTCTCGTGTTAAACTTAAATATCAACACATTATAACGAATAGTTTTGTGGAGTGCAACCGGCTGCTGCGCTGGTGCCCGTCCCCGGACTGCAGCAACGCGATCAAGGTGGCGTACGTGGAGGCGGCGCCGGTGACGTGCCGCTGCGGACACACGTTCTGCTTCGCCTGCGGAGAGAACTGGCACGATCCGGTCCGGTGCTGTTTACTGAGGAAGTGGATTAAGAAATGCGACGACGATTCGGAGACCTCAAATTGGATAGCGGCTAACACGAAGGAGTGTCCTAAATGCAACGTCACCATCGAGAAGGACGGCGGCTGCAACCACATGGTGTGCAAGAATCAGAATTGCAAGGCCGACTTCTGCTGGGTGTGTCTTGGACCTTGGGAGCCCCACGGCAGCAGCTGGTACAGCTGTAACAGATACGACGAGGACGAAGCGAAAGCAGCCCGGGACGCACAGGAGCGATCCCGCGCAGCGCTGCAGCGCTATCTGTTCTACTGCAACCGGTACATGAACCACATGCAGTCGCTGCGCTTCGAGTCCAAACTCTACGCCTCGGTTAAAGAGAAGATGGAGGAGATGCAGCAACACAACATGAGCTGGATTGAGGTCCAATTCCTCAAGAAAGCTGTAGATATTCTCTGCCAGTGCCGACAAACGCTGATGTACACGTACGTGTTCGCGTATTACTTGCGCAAGAACAACCAGTCGGTCATATTCGAGGACAACCAACGCGACTTGGAGTCGGCGACCGAAACGCTCTCGGAGTACTTGGAGCGGGACATCACTAGCGAGAACCTTGCCGACATCAAGCAGAAGGTTCAAGATAAATACAGATACTGTGACAGTCGGCGAAAAGTGTTATTAGAGCACGTGCACGAGGGATACGAGAAGGACTGCTGGGACTACACAGAGTAA
Protein
MDSEDDTKDDVDSGNESSGDDVDFAMDVETHSTRERQTELEEYPYEVLSTEEIVQHMVDCIKEVNTVVEIPATTTRILLNHFKWDKEKLMERFYDGDQDQLFSEARVINPFRKPVIQRPKLPRRISTSGMEECEICFSVLPTSIMTGLECGHRFCTQCWCEYLTTKIMEEGLGQTIACAAHACDILVDDATVMRLVRDSRVKLKYQHIITNSFVECNRLLRWCPSPDCSNAIKVAYVEAAPVTCRCGHTFCFACGENWHDPVRCCLLRKWIKKCDDDSETSNWIAANTKECPKCNVTIEKDGGCNHMVCKNQNCKADFCWVCLGPWEPHGSSWYSCNRYDEDEAKAARDAQERSRAALQRYLFYCNRYMNHMQSLRFESKLYASVKEKMEEMQQHNMSWIEVQFLKKAVDILCQCRQTLMYTYVFAYYLRKNNQSVIFEDNQRDLESATETLSEYLERDITSENLADIKQKVQDKYRYCDSRRKVLLEHVHEGYEKDCWDYTE
Summary
Catalytic Activity
[E2 ubiquitin-conjugating enzyme]-S-ubiquitinyl-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-N(6)-ubiquitinyl-L-lysine.
Similarity
Belongs to the RBR family.
Uniprot
H9IVY6
A0A3S2M918
A0A1E1WSH5
A0A2A4IZF6
A0A2A4IYG5
A0A1E1WJZ4
+ More
A0A194Q4U2 A0A2H1W2P6 A0A2J7R235 A0A067R7Y9 A0A224XMJ8 A0A023F5R3 A0A154P8K3 A0A0A9Y272 A0A0A9Y7R7 E0VRP1 T1IE78 A0A0A9Y4Q0 A0A310S672 A0A0A9YC94 A0A026W6K1 A0A0L7R9J6 A0A195C946 A0A151IWR7 A0A151WNB4 A0A2A4IY32 A0A195F4U3 A0A0N1ITV0 A0A2A3E4P3 A0A087ZQL8 V9I9T5 U4UWY5 E2ABE9 A0A1B6GNI4 A0A0J7NI82 A0A1B6G3Q4 E2B3Z6 J9JWN6 A0A2S2N673 A0A1Y1JR29 A0A0C9QQC7 A0A1B6HFM2 D6W822 A0A158N8X3 N6UI12 A0A2S2R1D4 A0A2P8YEJ4 A0A1B6DWQ4 K7IWP7 A0A232F279 A0A1B6DH29 A0A1Y1KMK1 A0A1B6DUY1 A0A1W4WL44 A0A1B6EDI8 E9IIR4 F4W9J2 U5EVD4 A0A1B0CHX7 A0A1L8DSV2 A0A0P6J528 A0A023EW10 A0A1Q3FFT3 N6TYD7 U4ULC7 A0A182LYG2 B0WDR8 A0A182WFY4 A0A182NC73 A0A1S4H580 A0A182KVN3 A0A182I5G2 A0A182WYD3 A0A182QQU8 A0A182RW30 A0A182YKK6 A0A2R5L864 A0A182JF80 A0A0T6BHD1 A0A2M4BL69 A0A2M4AA06 W5JVA5 A0A182F475 A0A182PJC8 A0A1Z5L7W4 A0A3R7NVW6 A0A2M3Z5U3 A0A0P4WLC4 A0A1Y9IUF0 A0A1Y9HDH3 L7MAF4 A0A1E1XQA3 A0A023GMZ8 G3MKV1 A0A1E1XCE1 A0A0N8DC36 A0A131XI44 A0A131Y0D8 B7P5K4 A0A0P6FQW6
A0A194Q4U2 A0A2H1W2P6 A0A2J7R235 A0A067R7Y9 A0A224XMJ8 A0A023F5R3 A0A154P8K3 A0A0A9Y272 A0A0A9Y7R7 E0VRP1 T1IE78 A0A0A9Y4Q0 A0A310S672 A0A0A9YC94 A0A026W6K1 A0A0L7R9J6 A0A195C946 A0A151IWR7 A0A151WNB4 A0A2A4IY32 A0A195F4U3 A0A0N1ITV0 A0A2A3E4P3 A0A087ZQL8 V9I9T5 U4UWY5 E2ABE9 A0A1B6GNI4 A0A0J7NI82 A0A1B6G3Q4 E2B3Z6 J9JWN6 A0A2S2N673 A0A1Y1JR29 A0A0C9QQC7 A0A1B6HFM2 D6W822 A0A158N8X3 N6UI12 A0A2S2R1D4 A0A2P8YEJ4 A0A1B6DWQ4 K7IWP7 A0A232F279 A0A1B6DH29 A0A1Y1KMK1 A0A1B6DUY1 A0A1W4WL44 A0A1B6EDI8 E9IIR4 F4W9J2 U5EVD4 A0A1B0CHX7 A0A1L8DSV2 A0A0P6J528 A0A023EW10 A0A1Q3FFT3 N6TYD7 U4ULC7 A0A182LYG2 B0WDR8 A0A182WFY4 A0A182NC73 A0A1S4H580 A0A182KVN3 A0A182I5G2 A0A182WYD3 A0A182QQU8 A0A182RW30 A0A182YKK6 A0A2R5L864 A0A182JF80 A0A0T6BHD1 A0A2M4BL69 A0A2M4AA06 W5JVA5 A0A182F475 A0A182PJC8 A0A1Z5L7W4 A0A3R7NVW6 A0A2M3Z5U3 A0A0P4WLC4 A0A1Y9IUF0 A0A1Y9HDH3 L7MAF4 A0A1E1XQA3 A0A023GMZ8 G3MKV1 A0A1E1XCE1 A0A0N8DC36 A0A131XI44 A0A131Y0D8 B7P5K4 A0A0P6FQW6
EC Number
2.3.2.31
Pubmed
19121390
26354079
24845553
25474469
25401762
26823975
+ More
20566863 24508170 30249741 23537049 20798317 28004739 18362917 19820115 21347285 29403074 20075255 28648823 21282665 21719571 26999592 24945155 12364791 20966253 25244985 20920257 23761445 28528879 25576852 29209593 22216098 28503490 28049606
20566863 24508170 30249741 23537049 20798317 28004739 18362917 19820115 21347285 29403074 20075255 28648823 21282665 21719571 26999592 24945155 12364791 20966253 25244985 20920257 23761445 28528879 25576852 29209593 22216098 28503490 28049606
EMBL
BABH01006368
RSAL01000006
RVE54291.1
GDQN01009458
GDQN01001126
JAT81596.1
+ More
JAT89928.1 NWSH01004601 PCG64866.1 PCG64865.1 GDQN01006980 GDQN01003730 JAT84074.1 JAT87324.1 KQ459585 KPI98425.1 ODYU01005827 SOQ47102.1 NEVH01008200 PNF34891.1 KK852643 KDR19556.1 GFTR01006766 JAW09660.1 GBBI01002197 JAC16515.1 KQ434844 KZC08183.1 GBHO01016482 GBRD01002612 JAG27122.1 JAG63209.1 GBHO01016481 GBRD01002611 GDHC01006969 JAG27123.1 JAG63210.1 JAQ11660.1 DS235478 EEB16047.1 ACPB03017568 GBHO01016480 GBRD01002610 JAG27124.1 JAG63211.1 KQ822452 OAD46915.1 GBHO01016479 GBRD01002609 GDHC01008448 JAG27125.1 JAG63212.1 JAQ10181.1 KK107372 QOIP01000004 EZA51742.1 RLU23234.1 KQ414624 KOC67542.1 KQ978068 KYM97389.1 KQ980849 KYN12223.1 KQ982907 KYQ49402.1 PCG64867.1 KQ981810 KYN35411.1 KQ435735 KOX77194.1 KZ288386 PBC26454.1 JR037590 JR037591 JR037592 AEY57858.1 AEY57859.1 AEY57860.1 KB632399 ERL94821.1 GL438237 EFN69270.1 GECZ01030188 GECZ01005763 GECZ01005292 JAS39581.1 JAS64006.1 JAS64477.1 LBMM01004640 KMQ92215.1 GECZ01028322 GECZ01012694 JAS41447.1 JAS57075.1 GL445428 EFN89583.1 ABLF02040829 GGMR01000028 MBY12647.1 GEZM01102975 GEZM01102974 JAV51654.1 GBYB01002837 GBYB01002839 JAG72604.1 JAG72606.1 GECU01034179 JAS73527.1 KQ971307 EFA11010.1 ADTU01008901 APGK01024787 APGK01024788 KB740562 ENN80276.1 GGMS01014377 MBY83580.1 PYGN01000659 PSN42682.1 GEDC01007196 JAS30102.1 NNAY01001253 OXU24609.1 GEDC01012299 JAS24999.1 GEZM01082016 JAV61440.1 GEDC01007802 JAS29496.1 GEDC01001331 JAS35967.1 GL763521 EFZ19528.1 GL888030 EGI69167.1 GANO01001081 JAB58790.1 AJWK01012844 AJWK01012845 AJWK01012846 AJWK01012847 GFDF01004555 JAV09529.1 GDUN01000710 JAN95209.1 GAPW01001079 JAC12519.1 GFDL01008637 JAV26408.1 APGK01050039 KB741165 ENN73396.1 KB632352 ERL93303.1 AXCM01007153 DS231900 EDS44821.1 AAAB01008823 APCN01003438 AXCN02001759 GGLE01001519 MBY05645.1 LJIG01000204 KRT86736.1 GGFJ01004603 MBW53744.1 GGFK01004127 MBW37448.1 ADMH02000248 ETN67248.1 GFJQ02003453 JAW03517.1 QCYY01002718 ROT68111.1 GGFM01003140 MBW23891.1 GDRN01039062 JAI67209.1 GACK01004911 JAA60123.1 GFAA01001942 JAU01493.1 GBBM01000895 JAC34523.1 JO842502 AEO34119.1 GFAC01002268 JAT96920.1 GDIQ01210403 GDIP01048359 LRGB01000024 JAK41322.1 JAM55356.1 KZS21710.1 GEFH01001778 JAP66803.1 GEFM01003844 JAP71952.1 ABJB010048594 ABJB010235798 ABJB010369897 ABJB010431572 ABJB010658312 ABJB010940894 ABJB011078547 DS641459 EEC01876.1 GDIQ01055565 JAN39172.1
JAT89928.1 NWSH01004601 PCG64866.1 PCG64865.1 GDQN01006980 GDQN01003730 JAT84074.1 JAT87324.1 KQ459585 KPI98425.1 ODYU01005827 SOQ47102.1 NEVH01008200 PNF34891.1 KK852643 KDR19556.1 GFTR01006766 JAW09660.1 GBBI01002197 JAC16515.1 KQ434844 KZC08183.1 GBHO01016482 GBRD01002612 JAG27122.1 JAG63209.1 GBHO01016481 GBRD01002611 GDHC01006969 JAG27123.1 JAG63210.1 JAQ11660.1 DS235478 EEB16047.1 ACPB03017568 GBHO01016480 GBRD01002610 JAG27124.1 JAG63211.1 KQ822452 OAD46915.1 GBHO01016479 GBRD01002609 GDHC01008448 JAG27125.1 JAG63212.1 JAQ10181.1 KK107372 QOIP01000004 EZA51742.1 RLU23234.1 KQ414624 KOC67542.1 KQ978068 KYM97389.1 KQ980849 KYN12223.1 KQ982907 KYQ49402.1 PCG64867.1 KQ981810 KYN35411.1 KQ435735 KOX77194.1 KZ288386 PBC26454.1 JR037590 JR037591 JR037592 AEY57858.1 AEY57859.1 AEY57860.1 KB632399 ERL94821.1 GL438237 EFN69270.1 GECZ01030188 GECZ01005763 GECZ01005292 JAS39581.1 JAS64006.1 JAS64477.1 LBMM01004640 KMQ92215.1 GECZ01028322 GECZ01012694 JAS41447.1 JAS57075.1 GL445428 EFN89583.1 ABLF02040829 GGMR01000028 MBY12647.1 GEZM01102975 GEZM01102974 JAV51654.1 GBYB01002837 GBYB01002839 JAG72604.1 JAG72606.1 GECU01034179 JAS73527.1 KQ971307 EFA11010.1 ADTU01008901 APGK01024787 APGK01024788 KB740562 ENN80276.1 GGMS01014377 MBY83580.1 PYGN01000659 PSN42682.1 GEDC01007196 JAS30102.1 NNAY01001253 OXU24609.1 GEDC01012299 JAS24999.1 GEZM01082016 JAV61440.1 GEDC01007802 JAS29496.1 GEDC01001331 JAS35967.1 GL763521 EFZ19528.1 GL888030 EGI69167.1 GANO01001081 JAB58790.1 AJWK01012844 AJWK01012845 AJWK01012846 AJWK01012847 GFDF01004555 JAV09529.1 GDUN01000710 JAN95209.1 GAPW01001079 JAC12519.1 GFDL01008637 JAV26408.1 APGK01050039 KB741165 ENN73396.1 KB632352 ERL93303.1 AXCM01007153 DS231900 EDS44821.1 AAAB01008823 APCN01003438 AXCN02001759 GGLE01001519 MBY05645.1 LJIG01000204 KRT86736.1 GGFJ01004603 MBW53744.1 GGFK01004127 MBW37448.1 ADMH02000248 ETN67248.1 GFJQ02003453 JAW03517.1 QCYY01002718 ROT68111.1 GGFM01003140 MBW23891.1 GDRN01039062 JAI67209.1 GACK01004911 JAA60123.1 GFAA01001942 JAU01493.1 GBBM01000895 JAC34523.1 JO842502 AEO34119.1 GFAC01002268 JAT96920.1 GDIQ01210403 GDIP01048359 LRGB01000024 JAK41322.1 JAM55356.1 KZS21710.1 GEFH01001778 JAP66803.1 GEFM01003844 JAP71952.1 ABJB010048594 ABJB010235798 ABJB010369897 ABJB010431572 ABJB010658312 ABJB010940894 ABJB011078547 DS641459 EEC01876.1 GDIQ01055565 JAN39172.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000235965
UP000027135
+ More
UP000076502 UP000009046 UP000015103 UP000053097 UP000279307 UP000053825 UP000078542 UP000078492 UP000075809 UP000078541 UP000053105 UP000242457 UP000005203 UP000030742 UP000000311 UP000036403 UP000008237 UP000007819 UP000007266 UP000005205 UP000019118 UP000245037 UP000002358 UP000215335 UP000192223 UP000007755 UP000092461 UP000075883 UP000002320 UP000075920 UP000075884 UP000075882 UP000075840 UP000076407 UP000075886 UP000075900 UP000076408 UP000075880 UP000000673 UP000069272 UP000075885 UP000283509 UP000076858 UP000001555
UP000076502 UP000009046 UP000015103 UP000053097 UP000279307 UP000053825 UP000078542 UP000078492 UP000075809 UP000078541 UP000053105 UP000242457 UP000005203 UP000030742 UP000000311 UP000036403 UP000008237 UP000007819 UP000007266 UP000005205 UP000019118 UP000245037 UP000002358 UP000215335 UP000192223 UP000007755 UP000092461 UP000075883 UP000002320 UP000075920 UP000075884 UP000075882 UP000075840 UP000076407 UP000075886 UP000075900 UP000076408 UP000075880 UP000000673 UP000069272 UP000075885 UP000283509 UP000076858 UP000001555
PRIDE
Interpro
IPR001841
Znf_RING
+ More
IPR031127 E3_UB_ligase_RBR
IPR002867 IBR_dom
IPR013083 Znf_RING/FYVE/PHD
IPR031126 Ariadne-2
IPR017907 Znf_RING_CS
IPR005937 26S_Psome_P45-like
IPR032501 Prot_ATP_ID_OB
IPR035263 PSMC6
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR003959 ATPase_AAA_core
IPR031127 E3_UB_ligase_RBR
IPR002867 IBR_dom
IPR013083 Znf_RING/FYVE/PHD
IPR031126 Ariadne-2
IPR017907 Znf_RING_CS
IPR005937 26S_Psome_P45-like
IPR032501 Prot_ATP_ID_OB
IPR035263 PSMC6
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR003959 ATPase_AAA_core
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
H9IVY6
A0A3S2M918
A0A1E1WSH5
A0A2A4IZF6
A0A2A4IYG5
A0A1E1WJZ4
+ More
A0A194Q4U2 A0A2H1W2P6 A0A2J7R235 A0A067R7Y9 A0A224XMJ8 A0A023F5R3 A0A154P8K3 A0A0A9Y272 A0A0A9Y7R7 E0VRP1 T1IE78 A0A0A9Y4Q0 A0A310S672 A0A0A9YC94 A0A026W6K1 A0A0L7R9J6 A0A195C946 A0A151IWR7 A0A151WNB4 A0A2A4IY32 A0A195F4U3 A0A0N1ITV0 A0A2A3E4P3 A0A087ZQL8 V9I9T5 U4UWY5 E2ABE9 A0A1B6GNI4 A0A0J7NI82 A0A1B6G3Q4 E2B3Z6 J9JWN6 A0A2S2N673 A0A1Y1JR29 A0A0C9QQC7 A0A1B6HFM2 D6W822 A0A158N8X3 N6UI12 A0A2S2R1D4 A0A2P8YEJ4 A0A1B6DWQ4 K7IWP7 A0A232F279 A0A1B6DH29 A0A1Y1KMK1 A0A1B6DUY1 A0A1W4WL44 A0A1B6EDI8 E9IIR4 F4W9J2 U5EVD4 A0A1B0CHX7 A0A1L8DSV2 A0A0P6J528 A0A023EW10 A0A1Q3FFT3 N6TYD7 U4ULC7 A0A182LYG2 B0WDR8 A0A182WFY4 A0A182NC73 A0A1S4H580 A0A182KVN3 A0A182I5G2 A0A182WYD3 A0A182QQU8 A0A182RW30 A0A182YKK6 A0A2R5L864 A0A182JF80 A0A0T6BHD1 A0A2M4BL69 A0A2M4AA06 W5JVA5 A0A182F475 A0A182PJC8 A0A1Z5L7W4 A0A3R7NVW6 A0A2M3Z5U3 A0A0P4WLC4 A0A1Y9IUF0 A0A1Y9HDH3 L7MAF4 A0A1E1XQA3 A0A023GMZ8 G3MKV1 A0A1E1XCE1 A0A0N8DC36 A0A131XI44 A0A131Y0D8 B7P5K4 A0A0P6FQW6
A0A194Q4U2 A0A2H1W2P6 A0A2J7R235 A0A067R7Y9 A0A224XMJ8 A0A023F5R3 A0A154P8K3 A0A0A9Y272 A0A0A9Y7R7 E0VRP1 T1IE78 A0A0A9Y4Q0 A0A310S672 A0A0A9YC94 A0A026W6K1 A0A0L7R9J6 A0A195C946 A0A151IWR7 A0A151WNB4 A0A2A4IY32 A0A195F4U3 A0A0N1ITV0 A0A2A3E4P3 A0A087ZQL8 V9I9T5 U4UWY5 E2ABE9 A0A1B6GNI4 A0A0J7NI82 A0A1B6G3Q4 E2B3Z6 J9JWN6 A0A2S2N673 A0A1Y1JR29 A0A0C9QQC7 A0A1B6HFM2 D6W822 A0A158N8X3 N6UI12 A0A2S2R1D4 A0A2P8YEJ4 A0A1B6DWQ4 K7IWP7 A0A232F279 A0A1B6DH29 A0A1Y1KMK1 A0A1B6DUY1 A0A1W4WL44 A0A1B6EDI8 E9IIR4 F4W9J2 U5EVD4 A0A1B0CHX7 A0A1L8DSV2 A0A0P6J528 A0A023EW10 A0A1Q3FFT3 N6TYD7 U4ULC7 A0A182LYG2 B0WDR8 A0A182WFY4 A0A182NC73 A0A1S4H580 A0A182KVN3 A0A182I5G2 A0A182WYD3 A0A182QQU8 A0A182RW30 A0A182YKK6 A0A2R5L864 A0A182JF80 A0A0T6BHD1 A0A2M4BL69 A0A2M4AA06 W5JVA5 A0A182F475 A0A182PJC8 A0A1Z5L7W4 A0A3R7NVW6 A0A2M3Z5U3 A0A0P4WLC4 A0A1Y9IUF0 A0A1Y9HDH3 L7MAF4 A0A1E1XQA3 A0A023GMZ8 G3MKV1 A0A1E1XCE1 A0A0N8DC36 A0A131XI44 A0A131Y0D8 B7P5K4 A0A0P6FQW6
PDB
4KC9
E-value=0,
Score=1890
Ontologies
GO
PANTHER
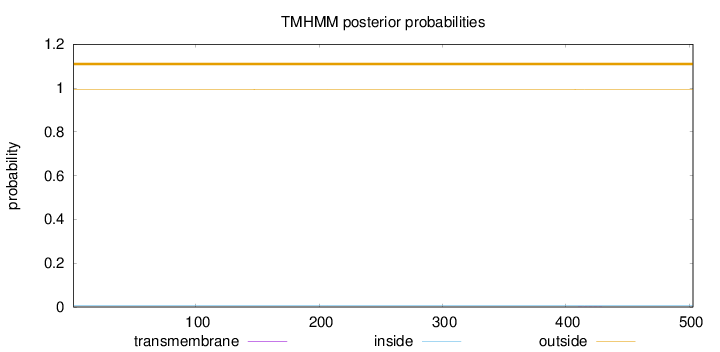
Topology
Length:
503
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0055
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00589
outside
1 - 503
Population Genetic Test Statistics
Pi
240.626378
Theta
178.7661
Tajima's D
0.553231
CLR
0.3275
CSRT
0.539523023848808
Interpretation
Uncertain
