Gene
KWMTBOMO12624
Pre Gene Modal
BGIBMGA001557
Annotation
para_sodium_channel_alpha_subunit_variant_2_[Bombyx_mandarina]
Full name
Sodium channel protein
Location in the cell
PlasmaMembrane Reliability : 4.278
Sequence
CDS
ATGTCCGAGGACTTGGACTCGATCAGCGAGGAAGAACGAAGCTTGTTCCGACCCTTCACACGAGAGTCTCTGGCCGCAATCGAAGCCCGCATAGCTGAAGAGCATGCCAAGCAAAAAGAACTCGAGAAAAAACGAGCGGAAGGCGAGAACGATTTGGGGCGGACGAAAAAGAAAAAAGAAGTGCGTTACGATGATGAGGATGAGGACGAGGGTCCCCAGCCGGACGCTACCTTGGAGCAAGGTCTACCACTGCCGGTGCGCATGCAAGGGTCCTTCCCGCTCGAACTCGCCTCTACACCACTCGAGGACATCGACCCCTTCTACCACAATCAGACAACCTTCGTAGTTATAAGCAAGGGTAAAGATATCTTCAGATTTTCGGCGACAAATGCTCTGTGGATATTAGACCCATTTAATCCCATAAGAAGAGTGGCTATATACATATTGGTACATCCTTTGTTTTCGCTGTTCATTATTACGACAATCTTAGTGAATTGCATACTTATGATAATGCCTACAACGCCAACAGTTGAAAGTACTGAAGTTATCTTTACCGGGATCTACACGTTTGAATCAGCGGTGAAAGTAATGGCCAGGGGTTTTATACTACAGCCATTCACATACCTTAGAGATGCATGGAATTGGCTTGACTTCGTAGTTATAGCTTTAGCTTATGTGACGATGGGCATAGATCTCGGAAACCTAGCCGCTCTGAGAACGTTCAGGGTACTACGAGCGTTGAAGACTGTGGCCATAGTACCGGGCTTGAAGACGATCGTCGGTGCTGTTATAGAATCCGTGAAAAATCTCCGTGATGTAATAATTTTAACGATGTTTTCGCTGTCCGTATTCGCGTTGATGGGCCTACAGATTTACATGGGAGTTTTGACGCAGAAGTGTGTCAAAGTGTTCCCGGAGGACGGCTCTTGGGGTAACCTGACCGACGAGAACTGGGAGAGGTTTTGTCAGAACGAAACTAACTGGTATGGTGAAGGGGGTGATTATCCTTTATGCGGCAATTCATCAGGGGCAGGACAATGCGAACCGGGCTACGTTTGTCTTCAAGGCTTCGGTCCAAATCCAAACTACGGATATACAAGCTTTGACACCTTCGGTTGGGCATTTCTATCAGCTTTCCGTCTAATGACACAGGATTATTGGGAAAATCTCTATCAATTGGTCTTAAGATCAGCTGGCTCGTGGCACGTATTGTTTTTCGTAGTGATAATATTCCTGGGATCTTTTTATCTGGTGAACTTGATTTTGGCTATCGTCGCCATGTCATACGATGAGCTACAGAAGAAAGCCGAAGAAGAAGAACAAGCAGAAGAAGAAGCTTTACGGGTACAGAATTAG
Protein
MSEDLDSISEEERSLFRPFTRESLAAIEARIAEEHAKQKELEKKRAEGENDLGRTKKKKEVRYDDEDEDEGPQPDATLEQGLPLPVRMQGSFPLELASTPLEDIDPFYHNQTTFVVISKGKDIFRFSATNALWILDPFNPIRRVAIYILVHPLFSLFIITTILVNCILMIMPTTPTVESTEVIFTGIYTFESAVKVMARGFILQPFTYLRDAWNWLDFVVIALAYVTMGIDLGNLAALRTFRVLRALKTVAIVPGLKTIVGAVIESVKNLRDVIILTMFSLSVFALMGLQIYMGVLTQKCVKVFPEDGSWGNLTDENWERFCQNETNWYGEGGDYPLCGNSSGAGQCEPGYVCLQGFGPNPNYGYTSFDTFGWAFLSAFRLMTQDYWENLYQLVLRSAGSWHVLFFVVIIFLGSFYLVNLILAIVAMSYDELQKKAEEEEQAEEEALRVQN
Summary
Description
Mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient.
Similarity
Belongs to the sodium channel (TC 1.A.1.10) family.
Uniprot
B3GEG0
B3GEF9
B3GEG1
B3GEF8
M4T8I8
A0A2H1W775
+ More
A0A1W6DWG8 A0A2A4J0G0 A0A1W6DWE6 Q94584 B3GEG2 B6SDJ2 C9D7C6 A0PAE8 A0PAE5 A0PAE6 A0PAE7 A0A212FAU8 A0A194Q2B4 A0A437BUZ8 A0A385I6P5 D7PI27 H9IWC6 A0A0D3R0X6 A0A0D3R104 A0A0D3R179 A0A2Z0LQW5 A0A240PKX8 A0A1W4XDT2 A0A1W4X4D2 A0A182NN78 A5I843 A0A1Y9HUC2 A0A2C9GUI5 A0A1W4XF20 A0A1S4GMH9 A0A1W4XF15 A0A1W4X4A1 A0A1W4X4D7 A0A1W4X4A6 A0A182QPX9 M1U7W8 A0A089ZWV3 A4IJ23 C3RTM3 C3RTM2 A0A182R2V3 A0A182WAV2 C3RTM1 A0A1W6S2I3 A0A1Z1EDE6 A0A2H4UZ54 A0A2H4UZ66 A0A1W6S2F2 B3VUH6 B3VUH5 A0A2H4UZ64 B3VUH7 D3KSS9 A0A2H4UZ65 A0A2H4UZ81 A0A2H4UZ84 A0A2H4UZ74 A0A2H4UZ92 A0A2H4UZ83 A0A2H4UZ77 A0A2H4UZ78 A0A2H4UZ99 A0A182IMT4 D3KST0 A0A2H4UZ93 C9D7C5 A0A232F412 K7IZT8 A0A3G4YHM1 A0A087ZV49 M1FNL3 A0A2R2WUN8 A0A0M8ZWZ2 K7W9N3 M1FIX3 C9D7C7 A1XNB4 M1FL24 A0A154PNY2 A0A182FLR9 A0A2K6VB92 A0A0L7RG34 C9D7C8 A0A1W4XDT8 A0A1W4XF25 A0A1W4XER0 A0A1B6LXE7 A0A1W4VUZ6 A0A1W4VVN8 A0A146LU77 A0A1S4GLX0 A0A0A9W7M5 A0A0Q9XNU3 A0A0Q9XEI2
A0A1W6DWG8 A0A2A4J0G0 A0A1W6DWE6 Q94584 B3GEG2 B6SDJ2 C9D7C6 A0PAE8 A0PAE5 A0PAE6 A0PAE7 A0A212FAU8 A0A194Q2B4 A0A437BUZ8 A0A385I6P5 D7PI27 H9IWC6 A0A0D3R0X6 A0A0D3R104 A0A0D3R179 A0A2Z0LQW5 A0A240PKX8 A0A1W4XDT2 A0A1W4X4D2 A0A182NN78 A5I843 A0A1Y9HUC2 A0A2C9GUI5 A0A1W4XF20 A0A1S4GMH9 A0A1W4XF15 A0A1W4X4A1 A0A1W4X4D7 A0A1W4X4A6 A0A182QPX9 M1U7W8 A0A089ZWV3 A4IJ23 C3RTM3 C3RTM2 A0A182R2V3 A0A182WAV2 C3RTM1 A0A1W6S2I3 A0A1Z1EDE6 A0A2H4UZ54 A0A2H4UZ66 A0A1W6S2F2 B3VUH6 B3VUH5 A0A2H4UZ64 B3VUH7 D3KSS9 A0A2H4UZ65 A0A2H4UZ81 A0A2H4UZ84 A0A2H4UZ74 A0A2H4UZ92 A0A2H4UZ83 A0A2H4UZ77 A0A2H4UZ78 A0A2H4UZ99 A0A182IMT4 D3KST0 A0A2H4UZ93 C9D7C5 A0A232F412 K7IZT8 A0A3G4YHM1 A0A087ZV49 M1FNL3 A0A2R2WUN8 A0A0M8ZWZ2 K7W9N3 M1FIX3 C9D7C7 A1XNB4 M1FL24 A0A154PNY2 A0A182FLR9 A0A2K6VB92 A0A0L7RG34 C9D7C8 A0A1W4XDT8 A0A1W4XF25 A0A1W4XER0 A0A1B6LXE7 A0A1W4VUZ6 A0A1W4VVN8 A0A146LU77 A0A1S4GLX0 A0A0A9W7M5 A0A0Q9XNU3 A0A0Q9XEI2
Pubmed
EMBL
EU688972
ACD80426.1
EU688971
ACD80425.1
EU688973
ACD80427.1
+ More
EU688970 ACD80424.1 KC342959 AGH70334.1 ODYU01006760 SOQ48897.1 KY247120 ARK07243.1 NWSH01004601 PCG64863.1 KY247121 ARK07244.1 AF072493 AH006308 AAC26517.1 EU688974 ACD80428.1 EU822499 ACJ09096.1 GQ202023 ACV87001.1 AB265178 BAF37096.2 AB265177 BAF37093.2 BAF37094.2 BAF37095.2 AGBW02009427 OWR50848.1 KQ459564 KPI99681.1 RSAL01000006 RVE54292.1 MG674159 AXY66618.1 GU574730 ADF80418.1 BABH01006366 BABH01006367 BABH01006368 KM027337 AJR27946.1 KM027335 AJR27944.1 KM027336 AJR27945.1 KJ699123 AJM87404.1 AM422833 CAM12801.1 AAAB01008968 AXCM01000653 AXCN02001896 KC107440 AGG53064.1 AB909019 BAP46858.1 BK005824 DAA05879.1 EU399181 ACB37024.1 EU399180 ACB37023.1 EU399179 KY747529 ACB37022.1 ARM35695.1 KY171977 ARO72115.1 KY747530 ARM35696.1 KY499805 AUA17903.1 KY499813 AUA17911.1 KY171978 ARO72116.1 EU817516 ACF04200.1 EU817515 ACF04199.1 KY499809 AUA17907.1 EU817517 ACF04201.1 AB453977 BAI77917.1 KY499808 KY499811 AUA17906.1 AUA17909.1 KY499806 AUA17904.1 KY499825 AUA17923.1 KY499818 AUA17916.1 KY499814 KY499817 AUA17912.1 AUA17915.1 KY499830 AUA17928.1 KY499822 AUA17920.1 KY499824 AUA17922.1 KY499827 AUA17925.1 AB453978 BAI77918.1 KY499826 AUA17924.1 GQ202022 KT455380 ACV87000.1 AMB38675.1 NNAY01001065 OXU25269.1 MG813771 AYV60955.1 JN695777 AFQ00696.1 KY123916 ARH02610.1 KQ435830 KOX71756.1 JX424548 AFW98420.1 JN695779 AFQ00698.1 GQ202024 ACV87002.1 DQ458470 ABE60888.1 JN695778 AFQ00697.1 KQ435007 KZC13595.1 KQ414598 KOC69810.1 GQ202025 ACV87003.1 GEBQ01011615 JAT28362.1 GDHC01008689 JAQ09940.1 GBHO01039810 JAG03794.1 CH933811 KRG06964.1 KRG06992.1
EU688970 ACD80424.1 KC342959 AGH70334.1 ODYU01006760 SOQ48897.1 KY247120 ARK07243.1 NWSH01004601 PCG64863.1 KY247121 ARK07244.1 AF072493 AH006308 AAC26517.1 EU688974 ACD80428.1 EU822499 ACJ09096.1 GQ202023 ACV87001.1 AB265178 BAF37096.2 AB265177 BAF37093.2 BAF37094.2 BAF37095.2 AGBW02009427 OWR50848.1 KQ459564 KPI99681.1 RSAL01000006 RVE54292.1 MG674159 AXY66618.1 GU574730 ADF80418.1 BABH01006366 BABH01006367 BABH01006368 KM027337 AJR27946.1 KM027335 AJR27944.1 KM027336 AJR27945.1 KJ699123 AJM87404.1 AM422833 CAM12801.1 AAAB01008968 AXCM01000653 AXCN02001896 KC107440 AGG53064.1 AB909019 BAP46858.1 BK005824 DAA05879.1 EU399181 ACB37024.1 EU399180 ACB37023.1 EU399179 KY747529 ACB37022.1 ARM35695.1 KY171977 ARO72115.1 KY747530 ARM35696.1 KY499805 AUA17903.1 KY499813 AUA17911.1 KY171978 ARO72116.1 EU817516 ACF04200.1 EU817515 ACF04199.1 KY499809 AUA17907.1 EU817517 ACF04201.1 AB453977 BAI77917.1 KY499808 KY499811 AUA17906.1 AUA17909.1 KY499806 AUA17904.1 KY499825 AUA17923.1 KY499818 AUA17916.1 KY499814 KY499817 AUA17912.1 AUA17915.1 KY499830 AUA17928.1 KY499822 AUA17920.1 KY499824 AUA17922.1 KY499827 AUA17925.1 AB453978 BAI77918.1 KY499826 AUA17924.1 GQ202022 KT455380 ACV87000.1 AMB38675.1 NNAY01001065 OXU25269.1 MG813771 AYV60955.1 JN695777 AFQ00696.1 KY123916 ARH02610.1 KQ435830 KOX71756.1 JX424548 AFW98420.1 JN695779 AFQ00698.1 GQ202024 ACV87002.1 DQ458470 ABE60888.1 JN695778 AFQ00697.1 KQ435007 KZC13595.1 KQ414598 KOC69810.1 GQ202025 ACV87003.1 GEBQ01011615 JAT28362.1 GDHC01008689 JAQ09940.1 GBHO01039810 JAG03794.1 CH933811 KRG06964.1 KRG06992.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
B3GEG0
B3GEF9
B3GEG1
B3GEF8
M4T8I8
A0A2H1W775
+ More
A0A1W6DWG8 A0A2A4J0G0 A0A1W6DWE6 Q94584 B3GEG2 B6SDJ2 C9D7C6 A0PAE8 A0PAE5 A0PAE6 A0PAE7 A0A212FAU8 A0A194Q2B4 A0A437BUZ8 A0A385I6P5 D7PI27 H9IWC6 A0A0D3R0X6 A0A0D3R104 A0A0D3R179 A0A2Z0LQW5 A0A240PKX8 A0A1W4XDT2 A0A1W4X4D2 A0A182NN78 A5I843 A0A1Y9HUC2 A0A2C9GUI5 A0A1W4XF20 A0A1S4GMH9 A0A1W4XF15 A0A1W4X4A1 A0A1W4X4D7 A0A1W4X4A6 A0A182QPX9 M1U7W8 A0A089ZWV3 A4IJ23 C3RTM3 C3RTM2 A0A182R2V3 A0A182WAV2 C3RTM1 A0A1W6S2I3 A0A1Z1EDE6 A0A2H4UZ54 A0A2H4UZ66 A0A1W6S2F2 B3VUH6 B3VUH5 A0A2H4UZ64 B3VUH7 D3KSS9 A0A2H4UZ65 A0A2H4UZ81 A0A2H4UZ84 A0A2H4UZ74 A0A2H4UZ92 A0A2H4UZ83 A0A2H4UZ77 A0A2H4UZ78 A0A2H4UZ99 A0A182IMT4 D3KST0 A0A2H4UZ93 C9D7C5 A0A232F412 K7IZT8 A0A3G4YHM1 A0A087ZV49 M1FNL3 A0A2R2WUN8 A0A0M8ZWZ2 K7W9N3 M1FIX3 C9D7C7 A1XNB4 M1FL24 A0A154PNY2 A0A182FLR9 A0A2K6VB92 A0A0L7RG34 C9D7C8 A0A1W4XDT8 A0A1W4XF25 A0A1W4XER0 A0A1B6LXE7 A0A1W4VUZ6 A0A1W4VVN8 A0A146LU77 A0A1S4GLX0 A0A0A9W7M5 A0A0Q9XNU3 A0A0Q9XEI2
A0A1W6DWG8 A0A2A4J0G0 A0A1W6DWE6 Q94584 B3GEG2 B6SDJ2 C9D7C6 A0PAE8 A0PAE5 A0PAE6 A0PAE7 A0A212FAU8 A0A194Q2B4 A0A437BUZ8 A0A385I6P5 D7PI27 H9IWC6 A0A0D3R0X6 A0A0D3R104 A0A0D3R179 A0A2Z0LQW5 A0A240PKX8 A0A1W4XDT2 A0A1W4X4D2 A0A182NN78 A5I843 A0A1Y9HUC2 A0A2C9GUI5 A0A1W4XF20 A0A1S4GMH9 A0A1W4XF15 A0A1W4X4A1 A0A1W4X4D7 A0A1W4X4A6 A0A182QPX9 M1U7W8 A0A089ZWV3 A4IJ23 C3RTM3 C3RTM2 A0A182R2V3 A0A182WAV2 C3RTM1 A0A1W6S2I3 A0A1Z1EDE6 A0A2H4UZ54 A0A2H4UZ66 A0A1W6S2F2 B3VUH6 B3VUH5 A0A2H4UZ64 B3VUH7 D3KSS9 A0A2H4UZ65 A0A2H4UZ81 A0A2H4UZ84 A0A2H4UZ74 A0A2H4UZ92 A0A2H4UZ83 A0A2H4UZ77 A0A2H4UZ78 A0A2H4UZ99 A0A182IMT4 D3KST0 A0A2H4UZ93 C9D7C5 A0A232F412 K7IZT8 A0A3G4YHM1 A0A087ZV49 M1FNL3 A0A2R2WUN8 A0A0M8ZWZ2 K7W9N3 M1FIX3 C9D7C7 A1XNB4 M1FL24 A0A154PNY2 A0A182FLR9 A0A2K6VB92 A0A0L7RG34 C9D7C8 A0A1W4XDT8 A0A1W4XF25 A0A1W4XER0 A0A1B6LXE7 A0A1W4VUZ6 A0A1W4VVN8 A0A146LU77 A0A1S4GLX0 A0A0A9W7M5 A0A0Q9XNU3 A0A0Q9XEI2
PDB
6A95
E-value=1.54928e-139,
Score=1272
Ontologies
GO
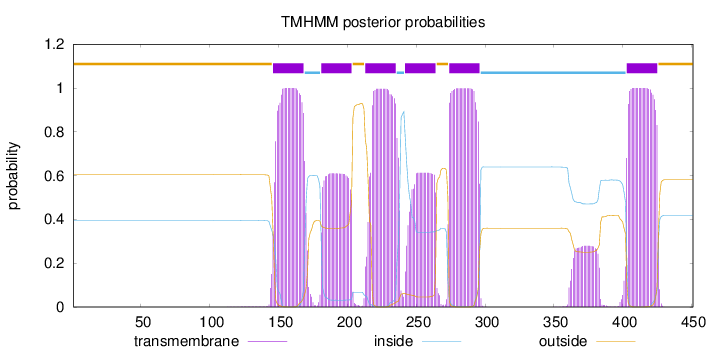
Topology
Subcellular location
Cell membrane
Length:
451
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
122.42922
Exp number, first 60 AAs:
0
Total prob of N-in:
0.39465
outside
1 - 145
TMhelix
146 - 168
inside
169 - 180
TMhelix
181 - 203
outside
204 - 212
TMhelix
213 - 235
inside
236 - 241
TMhelix
242 - 264
outside
265 - 273
TMhelix
274 - 296
inside
297 - 402
TMhelix
403 - 425
outside
426 - 451
Population Genetic Test Statistics
Pi
186.238437
Theta
180.832937
Tajima's D
-0.493077
CLR
2.068513
CSRT
0.243937803109845
Interpretation
Uncertain
