Gene
KWMTBOMO12616
Annotation
PREDICTED:_unconventional_myosin-IXa-like_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.127
Sequence
CDS
ATGCGAGAAGACGATATGAGTTCTAAGATCCGCGGCGTGCTGCAGGACCTGGCGACGCTGGAGACGGCGGCGCACACGGCGTCCAACCGGCTCTCCACGCTGCGCTCCACGAGCGCGCCGCAGCGACACGAGGAGCACATCCTCGTCGGACACATCAACGCCATCGAGAAGGAGAAGGCCATACTTACCAACGATCTGCCAACTCTCATCAGAGCCACATCAGACGACGACCTGTTGTCGACGACGGACCACGACGGCGAGCTCGGCAGCACGGACGACCTCAGCGCCACCTCGTCGCTGCGCTCGCAACGCCTCCTGCACCGGCAACTCTCCTCCGACGACCCCATCATGGTTTGA
Protein
MREDDMSSKIRGVLQDLATLETAAHTASNRLSTLRSTSAPQRHEEHILVGHINAIEKEKAILTNDLPTLIRATSDDDLLSTTDHDGELGSTDDLSATSSLRSQRLLHRQLSSDDPIMV
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
H9IVZ2
A0A2A4JC52
A0A194Q2A2
A0A194RE69
A0A3S2NR96
S4P5M9
+ More
A0A0K8TTD2 A0A1J1J244 A0A1B0GIC0 W4VR56 D6WCN7 A0A139WM08 A0A1W4WHT4 A0A1W4W7E8 A0A1W4WJ14 A0A1W4W7E6 A0A1W4WIS6 A0A1W4WHS4 A0A2S2Q9J5 A0A0A9ZHF6 E2B3B4 A0A2S2Q6E0 Q17D27 A0A154PE34 A0A182GTE4 A0A1S4G7K3 A0A1Y1K620 A0A1Y1K0Z7 A0A2J7QXG6 A0A067R2L8 A0A2J7QXH0 A0A2P8YLZ1 B0WP75 A0A182PUG0 A0A182F827 A0A182UKQ1 A0A182XH66 A0A182VXM2 A0A182RBE7 A0A3L8D8R1 A0A182K1V4 E2AQA0 Q7PMM4 A0A182JAL1 A0A1S4GJR6 A0A026WW74 A0A1I8JT52 W5J4I3 A0A1C7CZ94 A0A2M4CRR3 A0A0L7RFF9 A0A2M3Z0P2 A0A2M4CMH7 A0A182YRE1 A0A2A3E5W4 A0A087ZUJ1 A0A182M277 A0A195BUT3 A0A195CQZ6 A0A195FLM6 A0A151XIQ5 A0A182SVK0 A0A310SJM3 A0A182N6E9 A0A195E061 F4WRS2 A0A158NEN1 A0A182QNP2 K7IXK5 A0A232F4I6 A0A210QFS6 T1HV52 T1GBN4 U4UW94
A0A0K8TTD2 A0A1J1J244 A0A1B0GIC0 W4VR56 D6WCN7 A0A139WM08 A0A1W4WHT4 A0A1W4W7E8 A0A1W4WJ14 A0A1W4W7E6 A0A1W4WIS6 A0A1W4WHS4 A0A2S2Q9J5 A0A0A9ZHF6 E2B3B4 A0A2S2Q6E0 Q17D27 A0A154PE34 A0A182GTE4 A0A1S4G7K3 A0A1Y1K620 A0A1Y1K0Z7 A0A2J7QXG6 A0A067R2L8 A0A2J7QXH0 A0A2P8YLZ1 B0WP75 A0A182PUG0 A0A182F827 A0A182UKQ1 A0A182XH66 A0A182VXM2 A0A182RBE7 A0A3L8D8R1 A0A182K1V4 E2AQA0 Q7PMM4 A0A182JAL1 A0A1S4GJR6 A0A026WW74 A0A1I8JT52 W5J4I3 A0A1C7CZ94 A0A2M4CRR3 A0A0L7RFF9 A0A2M3Z0P2 A0A2M4CMH7 A0A182YRE1 A0A2A3E5W4 A0A087ZUJ1 A0A182M277 A0A195BUT3 A0A195CQZ6 A0A195FLM6 A0A151XIQ5 A0A182SVK0 A0A310SJM3 A0A182N6E9 A0A195E061 F4WRS2 A0A158NEN1 A0A182QNP2 K7IXK5 A0A232F4I6 A0A210QFS6 T1HV52 T1GBN4 U4UW94
Pubmed
EMBL
BABH01006359
BABH01006360
NWSH01002061
PCG69278.1
KQ459564
KPI99697.1
+ More
KQ460325 KPJ15764.1 RSAL01000006 RVE54297.1 GAIX01005424 JAA87136.1 GDAI01000006 JAI17597.1 CVRI01000064 CRL05526.1 AJWK01013646 GANO01004369 JAB55502.1 KQ971317 EEZ98803.1 KYB28980.1 GGMS01005216 MBY74419.1 GBHO01001939 JAG41665.1 GL445323 EFN89792.1 GGMS01003958 MBY73161.1 CH477301 EAT44232.1 KQ434886 KZC10136.1 JXUM01086865 JXUM01086866 JXUM01086867 JXUM01086868 JXUM01086869 JXUM01086870 JXUM01086871 JXUM01086872 KQ563596 KXJ73615.1 GEZM01096097 JAV55165.1 GEZM01096098 JAV55164.1 NEVH01009381 PNF33276.1 KK853055 KDR11981.1 PNF33279.1 PYGN01000501 PSN45275.1 DS232020 EDS32120.1 QOIP01000011 RLU16483.1 GL441701 EFN64434.1 AAAB01008978 EAA13585.6 KK107078 EZA60330.1 APCN01002667 APCN01002668 APCN01002669 APCN01002670 ADMH02002112 ETN58826.1 GGFL01003380 MBW67558.1 KQ414606 KOC69554.1 GGFM01001300 MBW22051.1 GGFL01002359 MBW66537.1 KZ288378 PBC26636.1 AXCM01008429 KQ976401 KYM92342.1 KQ977381 KYN03126.1 KQ981490 KYN41236.1 KQ982080 KYQ60284.1 KQ764228 OAD54572.1 KQ979999 KYN18264.1 GL888292 EGI63079.1 ADTU01013558 AXCN02000286 AAZX01005812 AAZX01011485 AAZX01023529 NNAY01000966 OXU25694.1 NEDP02003864 OWF47559.1 ACPB03023273 CAQQ02083912 KB632391 ERL94546.1
KQ460325 KPJ15764.1 RSAL01000006 RVE54297.1 GAIX01005424 JAA87136.1 GDAI01000006 JAI17597.1 CVRI01000064 CRL05526.1 AJWK01013646 GANO01004369 JAB55502.1 KQ971317 EEZ98803.1 KYB28980.1 GGMS01005216 MBY74419.1 GBHO01001939 JAG41665.1 GL445323 EFN89792.1 GGMS01003958 MBY73161.1 CH477301 EAT44232.1 KQ434886 KZC10136.1 JXUM01086865 JXUM01086866 JXUM01086867 JXUM01086868 JXUM01086869 JXUM01086870 JXUM01086871 JXUM01086872 KQ563596 KXJ73615.1 GEZM01096097 JAV55165.1 GEZM01096098 JAV55164.1 NEVH01009381 PNF33276.1 KK853055 KDR11981.1 PNF33279.1 PYGN01000501 PSN45275.1 DS232020 EDS32120.1 QOIP01000011 RLU16483.1 GL441701 EFN64434.1 AAAB01008978 EAA13585.6 KK107078 EZA60330.1 APCN01002667 APCN01002668 APCN01002669 APCN01002670 ADMH02002112 ETN58826.1 GGFL01003380 MBW67558.1 KQ414606 KOC69554.1 GGFM01001300 MBW22051.1 GGFL01002359 MBW66537.1 KZ288378 PBC26636.1 AXCM01008429 KQ976401 KYM92342.1 KQ977381 KYN03126.1 KQ981490 KYN41236.1 KQ982080 KYQ60284.1 KQ764228 OAD54572.1 KQ979999 KYN18264.1 GL888292 EGI63079.1 ADTU01013558 AXCN02000286 AAZX01005812 AAZX01011485 AAZX01023529 NNAY01000966 OXU25694.1 NEDP02003864 OWF47559.1 ACPB03023273 CAQQ02083912 KB632391 ERL94546.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000183832
+ More
UP000092461 UP000007266 UP000192223 UP000008237 UP000008820 UP000076502 UP000069940 UP000249989 UP000235965 UP000027135 UP000245037 UP000002320 UP000075885 UP000069272 UP000075902 UP000076407 UP000075920 UP000075900 UP000279307 UP000075881 UP000000311 UP000007062 UP000075880 UP000053097 UP000075840 UP000000673 UP000075882 UP000053825 UP000076408 UP000242457 UP000005203 UP000075883 UP000078540 UP000078542 UP000078541 UP000075809 UP000075901 UP000075884 UP000078492 UP000007755 UP000005205 UP000075886 UP000002358 UP000215335 UP000242188 UP000015103 UP000015102 UP000030742
UP000092461 UP000007266 UP000192223 UP000008237 UP000008820 UP000076502 UP000069940 UP000249989 UP000235965 UP000027135 UP000245037 UP000002320 UP000075885 UP000069272 UP000075902 UP000076407 UP000075920 UP000075900 UP000279307 UP000075881 UP000000311 UP000007062 UP000075880 UP000053097 UP000075840 UP000000673 UP000075882 UP000053825 UP000076408 UP000242457 UP000005203 UP000075883 UP000078540 UP000078542 UP000078541 UP000075809 UP000075901 UP000075884 UP000078492 UP000007755 UP000005205 UP000075886 UP000002358 UP000215335 UP000242188 UP000015103 UP000015102 UP000030742
PRIDE
Pfam
Interpro
IPR000048
IQ_motif_EF-hand-BS
+ More
IPR036023 MYSc_Myo9
IPR008936 Rho_GTPase_activation_prot
IPR000198 RhoGAP_dom
IPR001609 Myosin_head_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR000159 RA_dom
IPR002219 PE/DAG-bd
IPR029071 Ubiquitin-like_domsf
IPR027417 P-loop_NTPase
IPR013713 XPO2_central
IPR011989 ARM-like
IPR016024 ARM-type_fold
IPR005043 XPO2_C
IPR001494 Importin-beta_N
IPR022398 Peptidase_S8_His-AS
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR013783 Ig-like_fold
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR012337 RNaseH-like_sf
IPR002156 RNaseH_domain
IPR036397 RNaseH_sf
IPR036023 MYSc_Myo9
IPR008936 Rho_GTPase_activation_prot
IPR000198 RhoGAP_dom
IPR001609 Myosin_head_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR000159 RA_dom
IPR002219 PE/DAG-bd
IPR029071 Ubiquitin-like_domsf
IPR027417 P-loop_NTPase
IPR013713 XPO2_central
IPR011989 ARM-like
IPR016024 ARM-type_fold
IPR005043 XPO2_C
IPR001494 Importin-beta_N
IPR022398 Peptidase_S8_His-AS
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR013783 Ig-like_fold
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR012337 RNaseH-like_sf
IPR002156 RNaseH_domain
IPR036397 RNaseH_sf
SUPFAM
ProteinModelPortal
H9IVZ2
A0A2A4JC52
A0A194Q2A2
A0A194RE69
A0A3S2NR96
S4P5M9
+ More
A0A0K8TTD2 A0A1J1J244 A0A1B0GIC0 W4VR56 D6WCN7 A0A139WM08 A0A1W4WHT4 A0A1W4W7E8 A0A1W4WJ14 A0A1W4W7E6 A0A1W4WIS6 A0A1W4WHS4 A0A2S2Q9J5 A0A0A9ZHF6 E2B3B4 A0A2S2Q6E0 Q17D27 A0A154PE34 A0A182GTE4 A0A1S4G7K3 A0A1Y1K620 A0A1Y1K0Z7 A0A2J7QXG6 A0A067R2L8 A0A2J7QXH0 A0A2P8YLZ1 B0WP75 A0A182PUG0 A0A182F827 A0A182UKQ1 A0A182XH66 A0A182VXM2 A0A182RBE7 A0A3L8D8R1 A0A182K1V4 E2AQA0 Q7PMM4 A0A182JAL1 A0A1S4GJR6 A0A026WW74 A0A1I8JT52 W5J4I3 A0A1C7CZ94 A0A2M4CRR3 A0A0L7RFF9 A0A2M3Z0P2 A0A2M4CMH7 A0A182YRE1 A0A2A3E5W4 A0A087ZUJ1 A0A182M277 A0A195BUT3 A0A195CQZ6 A0A195FLM6 A0A151XIQ5 A0A182SVK0 A0A310SJM3 A0A182N6E9 A0A195E061 F4WRS2 A0A158NEN1 A0A182QNP2 K7IXK5 A0A232F4I6 A0A210QFS6 T1HV52 T1GBN4 U4UW94
A0A0K8TTD2 A0A1J1J244 A0A1B0GIC0 W4VR56 D6WCN7 A0A139WM08 A0A1W4WHT4 A0A1W4W7E8 A0A1W4WJ14 A0A1W4W7E6 A0A1W4WIS6 A0A1W4WHS4 A0A2S2Q9J5 A0A0A9ZHF6 E2B3B4 A0A2S2Q6E0 Q17D27 A0A154PE34 A0A182GTE4 A0A1S4G7K3 A0A1Y1K620 A0A1Y1K0Z7 A0A2J7QXG6 A0A067R2L8 A0A2J7QXH0 A0A2P8YLZ1 B0WP75 A0A182PUG0 A0A182F827 A0A182UKQ1 A0A182XH66 A0A182VXM2 A0A182RBE7 A0A3L8D8R1 A0A182K1V4 E2AQA0 Q7PMM4 A0A182JAL1 A0A1S4GJR6 A0A026WW74 A0A1I8JT52 W5J4I3 A0A1C7CZ94 A0A2M4CRR3 A0A0L7RFF9 A0A2M3Z0P2 A0A2M4CMH7 A0A182YRE1 A0A2A3E5W4 A0A087ZUJ1 A0A182M277 A0A195BUT3 A0A195CQZ6 A0A195FLM6 A0A151XIQ5 A0A182SVK0 A0A310SJM3 A0A182N6E9 A0A195E061 F4WRS2 A0A158NEN1 A0A182QNP2 K7IXK5 A0A232F4I6 A0A210QFS6 T1HV52 T1GBN4 U4UW94
Ontologies
GO
Topology
Subcellular location
Nucleus
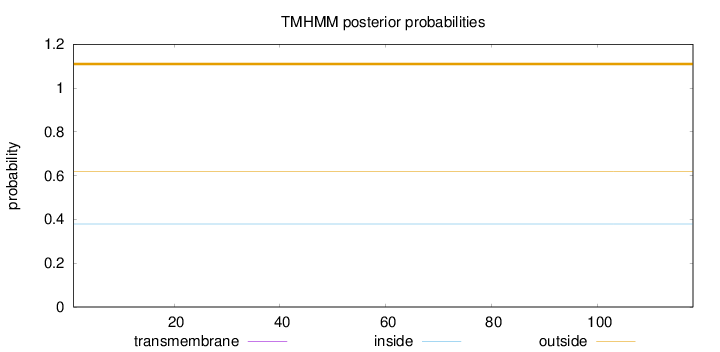
Length:
118
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0004
Exp number, first 60 AAs:
0.0004
Total prob of N-in:
0.37950
outside
1 - 118
Population Genetic Test Statistics
Pi
31.828486
Theta
52.981135
Tajima's D
-1.090816
CLR
0.548595
CSRT
0.127393630318484
Interpretation
Uncertain
