Gene
KWMTBOMO12613
Pre Gene Modal
BGIBMGA001552
Annotation
PREDICTED:_probable_phospholipid-transporting_ATPase_IM_isoform_X3_[Bombyx_mori]
Full name
Phospholipid-transporting ATPase
Location in the cell
PlasmaMembrane Reliability : 4.594
Sequence
CDS
ATGTGTGTCGATGGTGCGGCCGACATGGGAAAGTCCTCGGCGAGAACACATGGCCAAGAAAATGAACGAAGAATCAAAGCGAATAACAGGGAATATAACGCGCAGTTTCGTTATGCCAATAACTACATCAAGACTTCGAAGTATTCGATTATAACGTTCCTGCCGCTGAACTTGTTAGAACAGTTTCAGCGGCTAGCGAACTTTTATTTCTTGTGCCTGTTGGTCCTTCAACTGATCCCGGCTATCTCCTCGCTGACGCCGATAACCACAGCGATACCTCTTATTGGTGTGCTAGCCCTCACTGCCGTCAAGGACGCTTACGACGATTTCCAACGTCATCAAAACGATTCGCAAGTGAATCACAGGCGAGCCAAGACCTTGCGTAATGGCAAGCTTGTGGAGGAGAAATGGGCGTCTGTTCAGGTGGGAGATGTCATCAGGTTGGAGAACGACCAGTTCGTCGCGGCCGACATCCTACTTCTCAGCAGCTCGGAACCTAACGGACTTTGTTACATAGAAACCGCCGAGCTTGATGGGGAGACGAATTTGAAATGTCGTCAGTGCCTTTTGGAGACGGCCGCAATGGGCCAAGACGACGCACAACTGGGCGCGTTCGATGGTGAAATTGTTTGCGAAACGCCCAACAATCTACTTAATAAGTTTGAAGGTACGTTAAGTTGGCGTAACCAACACTTTTCACTCGACAACGATAAGATTCTGTTGCGCGGATGCGTGCTCCGCAACACGTCGTGGTGCTACGGCGTCGTTGTCTTTGCCGGGAAGGACACTAAACTAATGCAGAACTCCGGCAAGACGAAGTTCAAGCGCACCAGCATAGACAGACTACTTAATTTTCTTATCATAGGGATAGTATTCTTTCTGCTGTCTATGTGCGTGTTCTGCACGGTGGCGTGCGGGGTGTGGGAGTGGCTGGTGGGGCGCTACTTCCAGGTGTACCTGCCTTGGGACACGCTGGTGCCGGCCGACCCCACGTGGGGCGCTGTCATCATCGCCCTGCTCGTGTTCTTCTCTTACGCCATCGTGATGAACACTGTCGTGCCTATATCGCTTTATGTTTCTGTTGAAGTTATAAGATTTGCCCAGAGTTTTCTTATTAATTGGGATGAAAAAATGTACTATGAAAAAACTGGTACCGCAGCCAAAGCGCGGACCACCACACTTAACGAAGAACTAGGGCAAATCCAGTATATATTTTCTGATAAAACGGGAACTCTCACGCAAAACATAATGACGTTCAATAAGTGTTCGATAGCGGGCGTTTGCTACGGAGACGTCGTGGACGAGACCACCGGGGAGACTCTGGAGATAAACGAGAGCTCGGAGCCTTTAGATTTCTCGTTCAATCCTGAATACGAGCCGGAGTTTAAGTTCTTCGATCCGACGCTACTAGACGCGGTGAAGCGCGGGGATTCACACGTTCACGACTTCTTTCGTCTCTTGGCGCTTTGTCACACGGTCATGCCGGACCAGAAGAATGGACGGCTCGAATACCAGGCCCAATCGCCAGACGAGTCGGCGCTGGTGTCCGCAGCAAGAAACTTCGGTTTTGTGTTTAGAGAACGTTCGCCTAATAGTATTACAATAGAAGTAATGGGTAACACAGAGGTGTATGAATTGTTATGTATATTAGATTTTAATAATGTTAGAAAAAGAATGTCCGTGATATTGAGGAAGGATGGTGAAATTAGGTTATATACAAAAGGTGCTGATAATGTAATATATGAAAGATTAAAGGCAACTAAAGGAGATGTTAAGTCTAAAACACAGGAACATTTAAATAAATTCGCCGGGGAAGGTTTGAGGACACTGGCGCTGGCGTGGCGGCCGCTGGAGGAGCGCGGGTTCGCCGAGTGGAAGCGCCGCCACCACGCCGCTGCGCTCGCGCTGCACGACCGGGACGAGCAGCTCGACGCCATCTACGAGGAGATCGAGACCGACCTGCTACTGCTGGGCGTGACAGCCATTGAAGATAAACTACAAGATGGAGTACCAGAAACAATAGCGAATCTCAGCATGGCCGGTATAAAGATCTGGGTGTTAACAGGAGATAAACAAGAAACCGCTATAAACATCGGGTACTCGTGTCAGTTGCTGACCGACGACATGGCGGAGGTGTTCGTCGTCGACGGCGCGTCCTACGATGACGTCAACAAGCAGCTGGCCAAGTGCAGGGATTCGATACGGGTCGTCAACACCTTCATGCCGCACGGTCCCCGGTCGTCGGATGCGAAGCGCGACGAGGAGCCTAACGGGGCGGTGTCGGGCCGCGGCGCCAACGTCAAGCTGAACGCGCCCGCCGTCTCCGTCGTCACGTTTAGCAACGAGTACGTGTCTGGGGGCGCGCACACCTCTGAGCCTCACGAGCACGCCAACGACGACTCCAACGGGTTCGCCATCGTCGTCAACGGGCACTCGCTAGTGCACTGTCTGCACCCTAAACTAGAGGAAAAATTTTCAGAAGTAGTATTACACTGTCGATCTGTGATCTGTTGTCGAGTGACTCCACTGCAAAAGGCGATGGTGGTAGAATTAATAAAGAAAAGCAGAAAAGCCGTGACGCTGGCTATAGGCGACGGAGCCAACGACGTGTCCATGATCAAAGCGGCCCACATCGGCGTGGGCATATCAGGCCAAGAGGGCATGCAGGCGGTGCTCGCGTCGGACTACTCCATAGCCCAGTTCCGGTTCCTCCAGCGCCTATTGCTGGTGCACGGGCGCTGGTCCTACTACCGGATGTGTAAATTCCTAAGATATTTCTTCTACAAAAACTTCGCATTCACCGTCTGTCATTTCTGGTTCGCGTTCTTCTGTGGATTCAGTGCGCAGACGGTGTTCGACGAGATGTTCATCTCGGTGTACAACCTGTTCTACACGTCGCTGCCGGTGCTGGCGCTGGGCGTGTTCGAGCAGGACGTGTCGGACGCGACGTCGCTGCAGTTCCCGAAGCTGTACGCGCCGGGACACACCAGCCAGCTCTTCAACAAGACGGAGTTCATCAAGTCCACGCTGCACGGCTGCTTCACCTCGCTCGTGCTCTTCCTCATCCCTTACGGTACGTACAACGATGGGCTGGCGGCGGACGGGCGGGTGCTGTCGGACCACATGCTCCTGGGCAGCGTAGTGGCCACGATACTGATAATAGATAATACAACTCAGATTGCACTGGACACGACGTACTGGACCGTGTTCAACCACGTCACGATCTGGGGCTCGCTCGTGTCGTACTTCGTGCTGGACTACTTCTACAACTACGCCATCGGCGGCCCGTACGTCGGCTCGCTCACCGTGGCGCTCACGCAACCCACCTTCTGGTTCACGGCCGTATTGACCATGATAATCCTGATGGTGCCGGTGGTGTCGTGGCGGCTGGCGAGCGCCCGAGCGCGCGGCACGCTGGCGGAGCGCCTGCGGCTGCGGCAGCGGTGGCGCGGCGCCGCCCCCGCCCCCCGCACTCCCTCCGCCCGCCGCGCGCGCCGCTCCGTGCGCTCCGGGTACGCCTTCGCGCACCAGGAGGGCTTCGGCCGTCTCATCACGTCCGGCAAGATCATGCGCCGCATCCCGCACGCCGACGCCGCCTTCTCCGCGCTCGCCTCGCTGCCCGGGAAGTTCCACGCGCGCCCCTCCTCGCCGCCACCAGAGCCCAAGGCGCCCACGCAGAACCTTGATACAGTCGACCTGTGA
Protein
MCVDGAADMGKSSARTHGQENERRIKANNREYNAQFRYANNYIKTSKYSIITFLPLNLLEQFQRLANFYFLCLLVLQLIPAISSLTPITTAIPLIGVLALTAVKDAYDDFQRHQNDSQVNHRRAKTLRNGKLVEEKWASVQVGDVIRLENDQFVAADILLLSSSEPNGLCYIETAELDGETNLKCRQCLLETAAMGQDDAQLGAFDGEIVCETPNNLLNKFEGTLSWRNQHFSLDNDKILLRGCVLRNTSWCYGVVVFAGKDTKLMQNSGKTKFKRTSIDRLLNFLIIGIVFFLLSMCVFCTVACGVWEWLVGRYFQVYLPWDTLVPADPTWGAVIIALLVFFSYAIVMNTVVPISLYVSVEVIRFAQSFLINWDEKMYYEKTGTAAKARTTTLNEELGQIQYIFSDKTGTLTQNIMTFNKCSIAGVCYGDVVDETTGETLEINESSEPLDFSFNPEYEPEFKFFDPTLLDAVKRGDSHVHDFFRLLALCHTVMPDQKNGRLEYQAQSPDESALVSAARNFGFVFRERSPNSITIEVMGNTEVYELLCILDFNNVRKRMSVILRKDGEIRLYTKGADNVIYERLKATKGDVKSKTQEHLNKFAGEGLRTLALAWRPLEERGFAEWKRRHHAAALALHDRDEQLDAIYEEIETDLLLLGVTAIEDKLQDGVPETIANLSMAGIKIWVLTGDKQETAINIGYSCQLLTDDMAEVFVVDGASYDDVNKQLAKCRDSIRVVNTFMPHGPRSSDAKRDEEPNGAVSGRGANVKLNAPAVSVVTFSNEYVSGGAHTSEPHEHANDDSNGFAIVVNGHSLVHCLHPKLEEKFSEVVLHCRSVICCRVTPLQKAMVVELIKKSRKAVTLAIGDGANDVSMIKAAHIGVGISGQEGMQAVLASDYSIAQFRFLQRLLLVHGRWSYYRMCKFLRYFFYKNFAFTVCHFWFAFFCGFSAQTVFDEMFISVYNLFYTSLPVLALGVFEQDVSDATSLQFPKLYAPGHTSQLFNKTEFIKSTLHGCFTSLVLFLIPYGTYNDGLAADGRVLSDHMLLGSVVATILIIDNTTQIALDTTYWTVFNHVTIWGSLVSYFVLDYFYNYAIGGPYVGSLTVALTQPTFWFTAVLTMIILMVPVVSWRLASARARGTLAERLRLRQRWRGAAPAPRTPSARRARRSVRSGYAFAHQEGFGRLITSGKIMRRIPHADAAFSALASLPGKFHARPSSPPPEPKAPTQNLDTVDL
Summary
Catalytic Activity
ATP + H(2)O + phospholipid(Side 1) = ADP + phosphate + phospholipid(Side 2).
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IV subfamily.
Feature
chain Phospholipid-transporting ATPase
Uniprot
A0A3S2P7H2
A0A212FAS6
A0A1W4WGI1
A0A1W4W512
A0A1W4W4Z0
A0A182JP59
+ More
A0A182M6L0 A0A182IJ29 D6X506 A0A067QZ44 A0A1Y1LAE6 A0A182VPH0 A0A1Y1L8X4 A0A139WAP4 A0A1L8DDT4 A0A182LEE3 A0A182Q512 A0A182Y6N5 V5I8E4 Q7PQW3 W8B3M7 W8AZH5 A0A034V3Z4 A0A0A1X7E8 A0A1A9WIE1 W8BEM1 A0A182P3T7 A0A0L0C3I5 T1PJK4 A0A182U8J3 A0A1B0AFD3 A0A1I8M1V8 A0A182XAK3 A0A0P4VGW7 A0A224XI73 A0A069DXW4 A0A0V0G338 A0A1I8PSV1 A0A1J1HFS6 A0A0Q9WPN5 A0A0Q9WMU9 B4M1Y5 A0A0B4LH22 A0A0P8YDB6 B4NDR3 A0A0M4EXT8 A0A2H1VYH6 A0A0R1E730 A0A1W4UPT3 A0A0B4K669 A0A1W4UMX3 A0A0R3NIU5 E0VRM3 A0A146MAI4 A0A0A9XD29 A0A0N7ZAC6 A0A2J7PNZ2 A0A2J7PNX8 A0A2J7PNX9 A0A2R5LMC8 A0A1W7RAX2 A0A131YJ96 A0A0K8SBF1 T1JEU8 A0A0K8SBG7 A0A1D2N4D5 V5H2L6 T1K886 A0A443R225 A0A1E1XFF3 L7LUF6 V5H495 A0A3S1I231 A0A034V666 A0A210PEG7 A0A0K2U7W1 A0A1W0WRG2 A0A1S3IFH4 A0A1S3IF50 A0A016WML9 A0A016WMH0 A0A016WNL1 A0A158QRU1 U4PBV8 Q7YXV5 U4PBI0 U4PEQ6 U4PRL1 Q9TXV2 A0A3Q7PFF2 A0A3Q7N722 A0A3Q7N0W2 A0A085MC71 A0A085NQE4 A0A2G5TYQ7 A0A2G5TYN3 A0A0V1BHM2 A0A0V1BG57 A0A0V1KUD2
A0A182M6L0 A0A182IJ29 D6X506 A0A067QZ44 A0A1Y1LAE6 A0A182VPH0 A0A1Y1L8X4 A0A139WAP4 A0A1L8DDT4 A0A182LEE3 A0A182Q512 A0A182Y6N5 V5I8E4 Q7PQW3 W8B3M7 W8AZH5 A0A034V3Z4 A0A0A1X7E8 A0A1A9WIE1 W8BEM1 A0A182P3T7 A0A0L0C3I5 T1PJK4 A0A182U8J3 A0A1B0AFD3 A0A1I8M1V8 A0A182XAK3 A0A0P4VGW7 A0A224XI73 A0A069DXW4 A0A0V0G338 A0A1I8PSV1 A0A1J1HFS6 A0A0Q9WPN5 A0A0Q9WMU9 B4M1Y5 A0A0B4LH22 A0A0P8YDB6 B4NDR3 A0A0M4EXT8 A0A2H1VYH6 A0A0R1E730 A0A1W4UPT3 A0A0B4K669 A0A1W4UMX3 A0A0R3NIU5 E0VRM3 A0A146MAI4 A0A0A9XD29 A0A0N7ZAC6 A0A2J7PNZ2 A0A2J7PNX8 A0A2J7PNX9 A0A2R5LMC8 A0A1W7RAX2 A0A131YJ96 A0A0K8SBF1 T1JEU8 A0A0K8SBG7 A0A1D2N4D5 V5H2L6 T1K886 A0A443R225 A0A1E1XFF3 L7LUF6 V5H495 A0A3S1I231 A0A034V666 A0A210PEG7 A0A0K2U7W1 A0A1W0WRG2 A0A1S3IFH4 A0A1S3IF50 A0A016WML9 A0A016WMH0 A0A016WNL1 A0A158QRU1 U4PBV8 Q7YXV5 U4PBI0 U4PEQ6 U4PRL1 Q9TXV2 A0A3Q7PFF2 A0A3Q7N722 A0A3Q7N0W2 A0A085MC71 A0A085NQE4 A0A2G5TYQ7 A0A2G5TYN3 A0A0V1BHM2 A0A0V1BG57 A0A0V1KUD2
EC Number
7.6.2.1
Pubmed
22118469
18362917
19820115
24845553
28004739
20966253
+ More
25244985 12364791 24495485 25348373 25830018 26108605 25315136 27129103 26334808 17994087 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 15632085 20566863 26823975 25401762 26830274 27289101 25765539 28503490 25576852 28812685 25730766 9851916 24929829
25244985 12364791 24495485 25348373 25830018 26108605 25315136 27129103 26334808 17994087 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 15632085 20566863 26823975 25401762 26830274 27289101 25765539 28503490 25576852 28812685 25730766 9851916 24929829
EMBL
RSAL01000006
RVE54299.1
AGBW02009427
OWR50842.1
AXCM01002117
KQ971381
+ More
EEZ97214.2 KK853171 KDR10311.1 GEZM01066269 JAV68027.1 GEZM01066270 JAV68026.1 KYB24975.1 GFDF01009445 JAV04639.1 AXCN02001467 GALX01004830 JAB63636.1 AAAB01008859 EAA08202.6 GAMC01014902 JAB91653.1 GAMC01014903 JAB91652.1 GAKP01021748 JAC37204.1 GBXI01007068 JAD07224.1 GAMC01014904 JAB91651.1 JRES01001043 KNC26014.1 KA648285 AFP62914.1 GDKW01002998 JAI53597.1 GFTR01008685 JAW07741.1 GBGD01000159 JAC88730.1 GECL01003594 JAP02530.1 CVRI01000002 CRK86707.1 CH940651 KRF82341.1 KRF82340.1 EDW65689.2 KRF82339.1 AE014297 AHN57293.1 CH902617 KPU79470.1 CH964239 EDW81882.2 CP012528 ALC48655.1 ODYU01005171 SOQ45776.1 CM000160 KRK03508.1 AFH06386.1 CM000070 KRT00904.1 DS235478 EEB16029.1 GDHC01001858 JAQ16771.1 GBHO01025007 GDHC01016406 JAG18597.1 JAQ02223.1 GDRN01100874 JAI58472.1 NEVH01023281 PNF18041.1 PNF18044.1 PNF18043.1 GGLE01006546 MBY10672.1 GFAH01000089 JAV48300.1 GEDV01010411 JAP78146.1 GBRD01015209 JAG50617.1 JH432130 GBRD01015211 GBRD01015208 JAG50618.1 LJIJ01000231 ODN00134.1 GANP01007238 JAB77230.1 CAEY01001871 NCKU01002575 RWS09297.1 GFAC01001260 JAT97928.1 GACK01009343 JAA55691.1 GANP01012649 JAB71819.1 RQTK01000026 RUS90719.1 GAKP01021747 JAC37205.1 NEDP02076748 OWF34874.1 HACA01016784 CDW34145.1 MTYJ01000056 OQV17784.1 JARK01000193 EYC40856.1 EYC40855.1 EYC40857.1 UZAF01020393 VDO69474.1 BX284604 CDH93219.1 CCD72223.2 CDH93218.1 CDH93217.1 CDH93216.1 CCD72226.1 KL363204 KFD54817.1 KL367481 KFD71690.1 PDUG01000004 PIC32414.1 PIC32415.1 JYDH01000046 KRY36067.1 KRY36069.1 JYDW01000243 KRZ50984.1
EEZ97214.2 KK853171 KDR10311.1 GEZM01066269 JAV68027.1 GEZM01066270 JAV68026.1 KYB24975.1 GFDF01009445 JAV04639.1 AXCN02001467 GALX01004830 JAB63636.1 AAAB01008859 EAA08202.6 GAMC01014902 JAB91653.1 GAMC01014903 JAB91652.1 GAKP01021748 JAC37204.1 GBXI01007068 JAD07224.1 GAMC01014904 JAB91651.1 JRES01001043 KNC26014.1 KA648285 AFP62914.1 GDKW01002998 JAI53597.1 GFTR01008685 JAW07741.1 GBGD01000159 JAC88730.1 GECL01003594 JAP02530.1 CVRI01000002 CRK86707.1 CH940651 KRF82341.1 KRF82340.1 EDW65689.2 KRF82339.1 AE014297 AHN57293.1 CH902617 KPU79470.1 CH964239 EDW81882.2 CP012528 ALC48655.1 ODYU01005171 SOQ45776.1 CM000160 KRK03508.1 AFH06386.1 CM000070 KRT00904.1 DS235478 EEB16029.1 GDHC01001858 JAQ16771.1 GBHO01025007 GDHC01016406 JAG18597.1 JAQ02223.1 GDRN01100874 JAI58472.1 NEVH01023281 PNF18041.1 PNF18044.1 PNF18043.1 GGLE01006546 MBY10672.1 GFAH01000089 JAV48300.1 GEDV01010411 JAP78146.1 GBRD01015209 JAG50617.1 JH432130 GBRD01015211 GBRD01015208 JAG50618.1 LJIJ01000231 ODN00134.1 GANP01007238 JAB77230.1 CAEY01001871 NCKU01002575 RWS09297.1 GFAC01001260 JAT97928.1 GACK01009343 JAA55691.1 GANP01012649 JAB71819.1 RQTK01000026 RUS90719.1 GAKP01021747 JAC37205.1 NEDP02076748 OWF34874.1 HACA01016784 CDW34145.1 MTYJ01000056 OQV17784.1 JARK01000193 EYC40856.1 EYC40855.1 EYC40857.1 UZAF01020393 VDO69474.1 BX284604 CDH93219.1 CCD72223.2 CDH93218.1 CDH93217.1 CDH93216.1 CCD72226.1 KL363204 KFD54817.1 KL367481 KFD71690.1 PDUG01000004 PIC32414.1 PIC32415.1 JYDH01000046 KRY36067.1 KRY36069.1 JYDW01000243 KRZ50984.1
Proteomes
UP000283053
UP000007151
UP000192223
UP000075881
UP000075883
UP000075880
+ More
UP000007266 UP000027135 UP000075903 UP000075882 UP000075886 UP000076408 UP000007062 UP000091820 UP000075885 UP000037069 UP000075902 UP000092445 UP000095301 UP000076407 UP000095300 UP000183832 UP000008792 UP000000803 UP000007801 UP000007798 UP000092553 UP000002282 UP000192221 UP000001819 UP000009046 UP000235965 UP000094527 UP000015104 UP000285301 UP000271974 UP000242188 UP000085678 UP000024635 UP000038042 UP000268014 UP000001940 UP000286641 UP000230233 UP000054776 UP000054721
UP000007266 UP000027135 UP000075903 UP000075882 UP000075886 UP000076408 UP000007062 UP000091820 UP000075885 UP000037069 UP000075902 UP000092445 UP000095301 UP000076407 UP000095300 UP000183832 UP000008792 UP000000803 UP000007801 UP000007798 UP000092553 UP000002282 UP000192221 UP000001819 UP000009046 UP000235965 UP000094527 UP000015104 UP000285301 UP000271974 UP000242188 UP000085678 UP000024635 UP000038042 UP000268014 UP000001940 UP000286641 UP000230233 UP000054776 UP000054721
Interpro
IPR030348
ATP8B4
+ More
IPR032631 P-type_ATPase_N
IPR036412 HAD-like_sf
IPR023298 ATPase_P-typ_TM_dom_sf
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR001757 P_typ_ATPase
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006539 P-type_ATPase_IV
IPR032630 P_typ_ATPase_c
IPR018303 ATPase_P-typ_P_site
IPR023214 HAD_sf
IPR030350 FetA
IPR030347 ATP8B2
IPR032631 P-type_ATPase_N
IPR036412 HAD-like_sf
IPR023298 ATPase_P-typ_TM_dom_sf
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR001757 P_typ_ATPase
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006539 P-type_ATPase_IV
IPR032630 P_typ_ATPase_c
IPR018303 ATPase_P-typ_P_site
IPR023214 HAD_sf
IPR030350 FetA
IPR030347 ATP8B2
Gene 3D
ProteinModelPortal
A0A3S2P7H2
A0A212FAS6
A0A1W4WGI1
A0A1W4W512
A0A1W4W4Z0
A0A182JP59
+ More
A0A182M6L0 A0A182IJ29 D6X506 A0A067QZ44 A0A1Y1LAE6 A0A182VPH0 A0A1Y1L8X4 A0A139WAP4 A0A1L8DDT4 A0A182LEE3 A0A182Q512 A0A182Y6N5 V5I8E4 Q7PQW3 W8B3M7 W8AZH5 A0A034V3Z4 A0A0A1X7E8 A0A1A9WIE1 W8BEM1 A0A182P3T7 A0A0L0C3I5 T1PJK4 A0A182U8J3 A0A1B0AFD3 A0A1I8M1V8 A0A182XAK3 A0A0P4VGW7 A0A224XI73 A0A069DXW4 A0A0V0G338 A0A1I8PSV1 A0A1J1HFS6 A0A0Q9WPN5 A0A0Q9WMU9 B4M1Y5 A0A0B4LH22 A0A0P8YDB6 B4NDR3 A0A0M4EXT8 A0A2H1VYH6 A0A0R1E730 A0A1W4UPT3 A0A0B4K669 A0A1W4UMX3 A0A0R3NIU5 E0VRM3 A0A146MAI4 A0A0A9XD29 A0A0N7ZAC6 A0A2J7PNZ2 A0A2J7PNX8 A0A2J7PNX9 A0A2R5LMC8 A0A1W7RAX2 A0A131YJ96 A0A0K8SBF1 T1JEU8 A0A0K8SBG7 A0A1D2N4D5 V5H2L6 T1K886 A0A443R225 A0A1E1XFF3 L7LUF6 V5H495 A0A3S1I231 A0A034V666 A0A210PEG7 A0A0K2U7W1 A0A1W0WRG2 A0A1S3IFH4 A0A1S3IF50 A0A016WML9 A0A016WMH0 A0A016WNL1 A0A158QRU1 U4PBV8 Q7YXV5 U4PBI0 U4PEQ6 U4PRL1 Q9TXV2 A0A3Q7PFF2 A0A3Q7N722 A0A3Q7N0W2 A0A085MC71 A0A085NQE4 A0A2G5TYQ7 A0A2G5TYN3 A0A0V1BHM2 A0A0V1BG57 A0A0V1KUD2
A0A182M6L0 A0A182IJ29 D6X506 A0A067QZ44 A0A1Y1LAE6 A0A182VPH0 A0A1Y1L8X4 A0A139WAP4 A0A1L8DDT4 A0A182LEE3 A0A182Q512 A0A182Y6N5 V5I8E4 Q7PQW3 W8B3M7 W8AZH5 A0A034V3Z4 A0A0A1X7E8 A0A1A9WIE1 W8BEM1 A0A182P3T7 A0A0L0C3I5 T1PJK4 A0A182U8J3 A0A1B0AFD3 A0A1I8M1V8 A0A182XAK3 A0A0P4VGW7 A0A224XI73 A0A069DXW4 A0A0V0G338 A0A1I8PSV1 A0A1J1HFS6 A0A0Q9WPN5 A0A0Q9WMU9 B4M1Y5 A0A0B4LH22 A0A0P8YDB6 B4NDR3 A0A0M4EXT8 A0A2H1VYH6 A0A0R1E730 A0A1W4UPT3 A0A0B4K669 A0A1W4UMX3 A0A0R3NIU5 E0VRM3 A0A146MAI4 A0A0A9XD29 A0A0N7ZAC6 A0A2J7PNZ2 A0A2J7PNX8 A0A2J7PNX9 A0A2R5LMC8 A0A1W7RAX2 A0A131YJ96 A0A0K8SBF1 T1JEU8 A0A0K8SBG7 A0A1D2N4D5 V5H2L6 T1K886 A0A443R225 A0A1E1XFF3 L7LUF6 V5H495 A0A3S1I231 A0A034V666 A0A210PEG7 A0A0K2U7W1 A0A1W0WRG2 A0A1S3IFH4 A0A1S3IF50 A0A016WML9 A0A016WMH0 A0A016WNL1 A0A158QRU1 U4PBV8 Q7YXV5 U4PBI0 U4PEQ6 U4PRL1 Q9TXV2 A0A3Q7PFF2 A0A3Q7N722 A0A3Q7N0W2 A0A085MC71 A0A085NQE4 A0A2G5TYQ7 A0A2G5TYN3 A0A0V1BHM2 A0A0V1BG57 A0A0V1KUD2
PDB
6A69
E-value=1.0989e-11,
Score=174
Ontologies
GO
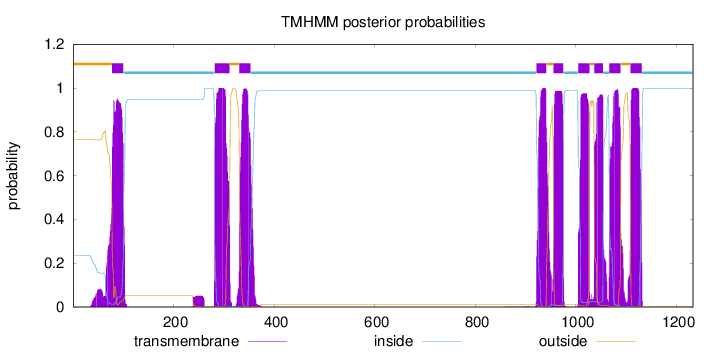
Topology
Subcellular location
Membrane
Length:
1233
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
203.38343
Exp number, first 60 AAs:
1.43746
Total prob of N-in:
0.23458
outside
1 - 77
TMhelix
78 - 100
inside
101 - 281
TMhelix
282 - 311
outside
312 - 330
TMhelix
331 - 353
inside
354 - 921
TMhelix
922 - 941
outside
942 - 955
TMhelix
956 - 975
inside
976 - 1004
TMhelix
1005 - 1027
outside
1028 - 1036
TMhelix
1037 - 1054
inside
1055 - 1066
TMhelix
1067 - 1089
outside
1090 - 1108
TMhelix
1109 - 1131
inside
1132 - 1233
Population Genetic Test Statistics
Pi
196.092795
Theta
17.68711
Tajima's D
-1.102085
CLR
0.508612
CSRT
0.12094395280236
Interpretation
Uncertain
