Gene
KWMTBOMO12606
Annotation
hypothetical_protein_RR46_09637_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 2.292
Sequence
CDS
ATGACAACAGGATTTACAGCTCGCGTAGACAACATACGAAATACATTAATTGCCGTAAGCAAGCGAAGAACTTTTAGATATGGTGTTCCGTTTATATTATTTGTTGTAGGAGGATCGTTTGGTTTGAGGGAATGGACACAAATTAGATATCAGTTTTCACAAGTAAAAGGTGTGAGTAAGGAAGAAGCAGAGAAAATGGGCCTTCACAGAGAAAAAACTGTTTCTTTGGATACTGCATATGAAGAAATACAAAAATTGGATATTGATAATTGGGAAAACAAACGAGGCCCTAGACCATGGGAAATAAATGAATCAAAAAAACACAATTAA
Protein
MTTGFTARVDNIRNTLIAVSKRRTFRYGVPFILFVVGGSFGLREWTQIRYQFSQVKGVSKEEAEKMGLHREKTVSLDTAYEEIQKLDIDNWENKRGPRPWEINESKKHN
Summary
Uniprot
A0A2A4JRE3
A0A3S2NLY0
A0A2H1VGS0
A0A194PYL2
A0A0L7KUL0
A0A182IXG2
+ More
A0A084VJB5 A0A182RNV2 A0A2P8YRS1 A0A182QXE5 A0A182N677 A0A067RPV2 A0A182PKP3 A0A182T987 A0A182Y470 D2A5N8 A0A1L8E127 A0A2M3ZKZ9 A0A1L8EBN4 A0A2M4C4A0 B3MX82 W5JHW8 A0A0K8TQ78 A0A182LYC1 A0A182JQ03 A0A182VHA3 A0A182TFI2 A0A182WRN7 Q7QD68 A0A182HU42 A0A182FRB4 A0A2M4AZC8 A0A023EEW8 A0A1I8PVB6 A0A1L8E9D5 A0A0T6B100 B4LZY6 N6U9S5 A0A0A9WSI4 A0A0P4VR50 R4FMS5 A0A0L0CPQ9 B4K5U7 B4NIS5 A0A1J1HX72 B4PS59 A0A0K8UJN7 B0W950 B3P0T8 A0A1I8M3C4 A0A293LFH9 R4WE56 A0A0K8R4F8 B7P5Y1 A0A2R7WBA6 A0A034WTF0 A0A1B0D933 A0A0C9RP53 A0A0V0G777 V5HXY5 Q177G5 A0A1Q3FCB4 A0A1W4VVF8 Q29QH7 Q294B1 A0A1B2AKH8 B4QZ20 A0A3B0JSE3 A0A069DVF8 A0A0K8WD33 B4HDY7 A0A2R5LD34 A0A1B6FN20 A0A336LHM2 A0A2R5LD22 A0A1Z5L8S7 A0A1A9W720 A0A131XQF3 A0A1E1WYB7 A0A023G0W6 T1J3W1 A0A0P4XBK8 A0A0P5FXU6 A0A1S3CXS7 G3MTB5 G3MTF2 A0A023G9G4 A0A1A9UZ38 A0A1B6C1Q2 A0A087TZP7 H3AVT1 A0A1B6DCY2 A0A023FPB7 A0A1B0G090 A0A151IEN1 A0A195FBQ1 A0A3S1BZY7
A0A084VJB5 A0A182RNV2 A0A2P8YRS1 A0A182QXE5 A0A182N677 A0A067RPV2 A0A182PKP3 A0A182T987 A0A182Y470 D2A5N8 A0A1L8E127 A0A2M3ZKZ9 A0A1L8EBN4 A0A2M4C4A0 B3MX82 W5JHW8 A0A0K8TQ78 A0A182LYC1 A0A182JQ03 A0A182VHA3 A0A182TFI2 A0A182WRN7 Q7QD68 A0A182HU42 A0A182FRB4 A0A2M4AZC8 A0A023EEW8 A0A1I8PVB6 A0A1L8E9D5 A0A0T6B100 B4LZY6 N6U9S5 A0A0A9WSI4 A0A0P4VR50 R4FMS5 A0A0L0CPQ9 B4K5U7 B4NIS5 A0A1J1HX72 B4PS59 A0A0K8UJN7 B0W950 B3P0T8 A0A1I8M3C4 A0A293LFH9 R4WE56 A0A0K8R4F8 B7P5Y1 A0A2R7WBA6 A0A034WTF0 A0A1B0D933 A0A0C9RP53 A0A0V0G777 V5HXY5 Q177G5 A0A1Q3FCB4 A0A1W4VVF8 Q29QH7 Q294B1 A0A1B2AKH8 B4QZ20 A0A3B0JSE3 A0A069DVF8 A0A0K8WD33 B4HDY7 A0A2R5LD34 A0A1B6FN20 A0A336LHM2 A0A2R5LD22 A0A1Z5L8S7 A0A1A9W720 A0A131XQF3 A0A1E1WYB7 A0A023G0W6 T1J3W1 A0A0P4XBK8 A0A0P5FXU6 A0A1S3CXS7 G3MTB5 G3MTF2 A0A023G9G4 A0A1A9UZ38 A0A1B6C1Q2 A0A087TZP7 H3AVT1 A0A1B6DCY2 A0A023FPB7 A0A1B0G090 A0A151IEN1 A0A195FBQ1 A0A3S1BZY7
Pubmed
26354079
26227816
24438588
29403074
24845553
25244985
+ More
18362917 19820115 17994087 20920257 23761445 26369729 12364791 14747013 17210077 24945155 18057021 23537049 25401762 26823975 27129103 26108605 17550304 25315136 23691247 25348373 25765539 17510324 15632085 26334808 28528879 28049606 28503490 22216098 9215903
18362917 19820115 17994087 20920257 23761445 26369729 12364791 14747013 17210077 24945155 18057021 23537049 25401762 26823975 27129103 26108605 17550304 25315136 23691247 25348373 25765539 17510324 15632085 26334808 28528879 28049606 28503490 22216098 9215903
EMBL
NWSH01000780
PCG74278.1
RSAL01000006
RVE54309.1
ODYU01002496
SOQ40049.1
+ More
KQ459585 KPI98421.1 JTDY01005635 KOB66800.1 ATLV01013502 KE524866 KFB38059.1 PYGN01000403 PSN46948.1 AXCN02000270 KK852541 KDR21754.1 KQ971345 EFA05396.1 GFDF01001683 JAV12401.1 GGFM01008421 MBW29172.1 GFDG01002767 JAV16032.1 GGFJ01010948 MBW60089.1 CH902629 EDV35305.1 ADMH02001204 ETN63716.1 GDAI01001558 JAI16045.1 AXCM01000193 AAAB01008859 EAA07588.3 APCN01000605 GGFK01012823 MBW46144.1 GAPW01006349 GAPW01006348 GEHC01000220 JAC07250.1 JAV47425.1 GFDG01003462 JAV15337.1 LJIG01016284 KRT81062.1 CH940650 EDW67214.2 KRF83184.1 APGK01043657 KB741015 KB632237 ENN75362.1 ERL90450.1 GBHO01034171 GBRD01004772 GDHC01018838 JAG09433.1 JAG61049.1 JAP99790.1 GDKW01001702 JAI54893.1 ACPB03016751 GAHY01001609 JAA75901.1 JRES01000091 KNC34167.1 CH933806 EDW15159.1 CH964272 EDW83789.2 CVRI01000024 CRK92122.1 CM000160 EDW97479.1 GDHF01025744 JAI26570.1 DS231862 EDS39747.1 CH954181 EDV48986.1 GFWV01001898 MAA26628.1 AK418119 BAN21334.1 GADI01007867 JAA65941.1 ABJB010228205 ABJB010506828 ABJB010789732 DS643033 EEC02003.1 KK854571 PTY16977.1 GAKP01000137 GAKP01000136 JAC58816.1 AJVK01012889 GBYB01010205 JAG79972.1 GECL01002283 JAP03841.1 GANP01003126 JAB81342.1 CH477376 EAT42312.1 GFDL01009829 JAV25216.1 BT024413 ABC86475.1 CM000070 EAL29053.2 KX531876 ANY27686.1 CM000364 EDX12848.1 OUUW01000008 SPP83923.1 GBGD01003590 JAC85299.1 GDHF01003216 JAI49098.1 CH480815 EDW42079.1 GGLE01003253 MBY07379.1 GECZ01018366 JAS51403.1 UFQS01000006 UFQT01000006 SSW96979.1 SSX17366.1 GGLE01003252 MBY07378.1 GFJQ02003536 JAW03434.1 GEFH01000261 JAP68320.1 GFAC01007169 JAT92019.1 GBBL01000810 JAC26510.1 JH431832 GDIP01243527 JAI79874.1 GDIQ01255993 JAJ95731.1 JO845116 AEO36733.1 JO845153 AEO36770.1 GBBM01005920 JAC29498.1 GEDC01030118 JAS07180.1 KK117482 KFM70586.1 AFYH01152278 AFYH01152279 GEDC01016822 GEDC01013843 JAS20476.1 JAS23455.1 GBBK01001348 JAC23134.1 CCAG010007751 KQ977862 KYM99280.1 KQ981693 KYN37811.1 RQTK01000463 RUS79237.1
KQ459585 KPI98421.1 JTDY01005635 KOB66800.1 ATLV01013502 KE524866 KFB38059.1 PYGN01000403 PSN46948.1 AXCN02000270 KK852541 KDR21754.1 KQ971345 EFA05396.1 GFDF01001683 JAV12401.1 GGFM01008421 MBW29172.1 GFDG01002767 JAV16032.1 GGFJ01010948 MBW60089.1 CH902629 EDV35305.1 ADMH02001204 ETN63716.1 GDAI01001558 JAI16045.1 AXCM01000193 AAAB01008859 EAA07588.3 APCN01000605 GGFK01012823 MBW46144.1 GAPW01006349 GAPW01006348 GEHC01000220 JAC07250.1 JAV47425.1 GFDG01003462 JAV15337.1 LJIG01016284 KRT81062.1 CH940650 EDW67214.2 KRF83184.1 APGK01043657 KB741015 KB632237 ENN75362.1 ERL90450.1 GBHO01034171 GBRD01004772 GDHC01018838 JAG09433.1 JAG61049.1 JAP99790.1 GDKW01001702 JAI54893.1 ACPB03016751 GAHY01001609 JAA75901.1 JRES01000091 KNC34167.1 CH933806 EDW15159.1 CH964272 EDW83789.2 CVRI01000024 CRK92122.1 CM000160 EDW97479.1 GDHF01025744 JAI26570.1 DS231862 EDS39747.1 CH954181 EDV48986.1 GFWV01001898 MAA26628.1 AK418119 BAN21334.1 GADI01007867 JAA65941.1 ABJB010228205 ABJB010506828 ABJB010789732 DS643033 EEC02003.1 KK854571 PTY16977.1 GAKP01000137 GAKP01000136 JAC58816.1 AJVK01012889 GBYB01010205 JAG79972.1 GECL01002283 JAP03841.1 GANP01003126 JAB81342.1 CH477376 EAT42312.1 GFDL01009829 JAV25216.1 BT024413 ABC86475.1 CM000070 EAL29053.2 KX531876 ANY27686.1 CM000364 EDX12848.1 OUUW01000008 SPP83923.1 GBGD01003590 JAC85299.1 GDHF01003216 JAI49098.1 CH480815 EDW42079.1 GGLE01003253 MBY07379.1 GECZ01018366 JAS51403.1 UFQS01000006 UFQT01000006 SSW96979.1 SSX17366.1 GGLE01003252 MBY07378.1 GFJQ02003536 JAW03434.1 GEFH01000261 JAP68320.1 GFAC01007169 JAT92019.1 GBBL01000810 JAC26510.1 JH431832 GDIP01243527 JAI79874.1 GDIQ01255993 JAJ95731.1 JO845116 AEO36733.1 JO845153 AEO36770.1 GBBM01005920 JAC29498.1 GEDC01030118 JAS07180.1 KK117482 KFM70586.1 AFYH01152278 AFYH01152279 GEDC01016822 GEDC01013843 JAS20476.1 JAS23455.1 GBBK01001348 JAC23134.1 CCAG010007751 KQ977862 KYM99280.1 KQ981693 KYN37811.1 RQTK01000463 RUS79237.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000037510
UP000075880
UP000030765
+ More
UP000075900 UP000245037 UP000075886 UP000075884 UP000027135 UP000075885 UP000075901 UP000076408 UP000007266 UP000007801 UP000000673 UP000075883 UP000075881 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000069272 UP000095300 UP000008792 UP000019118 UP000030742 UP000015103 UP000037069 UP000009192 UP000007798 UP000183832 UP000002282 UP000002320 UP000008711 UP000095301 UP000001555 UP000092462 UP000008820 UP000192221 UP000001819 UP000000304 UP000268350 UP000001292 UP000091820 UP000079169 UP000078200 UP000054359 UP000008672 UP000092444 UP000078542 UP000078541 UP000271974
UP000075900 UP000245037 UP000075886 UP000075884 UP000027135 UP000075885 UP000075901 UP000076408 UP000007266 UP000007801 UP000000673 UP000075883 UP000075881 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000069272 UP000095300 UP000008792 UP000019118 UP000030742 UP000015103 UP000037069 UP000009192 UP000007798 UP000183832 UP000002282 UP000002320 UP000008711 UP000095301 UP000001555 UP000092462 UP000008820 UP000192221 UP000001819 UP000000304 UP000268350 UP000001292 UP000091820 UP000079169 UP000078200 UP000054359 UP000008672 UP000092444 UP000078542 UP000078541 UP000271974
PRIDE
Pfam
PF14138 COX16
Interpro
IPR020164
Cyt_c_Oxase_assmbl_COX16
ProteinModelPortal
A0A2A4JRE3
A0A3S2NLY0
A0A2H1VGS0
A0A194PYL2
A0A0L7KUL0
A0A182IXG2
+ More
A0A084VJB5 A0A182RNV2 A0A2P8YRS1 A0A182QXE5 A0A182N677 A0A067RPV2 A0A182PKP3 A0A182T987 A0A182Y470 D2A5N8 A0A1L8E127 A0A2M3ZKZ9 A0A1L8EBN4 A0A2M4C4A0 B3MX82 W5JHW8 A0A0K8TQ78 A0A182LYC1 A0A182JQ03 A0A182VHA3 A0A182TFI2 A0A182WRN7 Q7QD68 A0A182HU42 A0A182FRB4 A0A2M4AZC8 A0A023EEW8 A0A1I8PVB6 A0A1L8E9D5 A0A0T6B100 B4LZY6 N6U9S5 A0A0A9WSI4 A0A0P4VR50 R4FMS5 A0A0L0CPQ9 B4K5U7 B4NIS5 A0A1J1HX72 B4PS59 A0A0K8UJN7 B0W950 B3P0T8 A0A1I8M3C4 A0A293LFH9 R4WE56 A0A0K8R4F8 B7P5Y1 A0A2R7WBA6 A0A034WTF0 A0A1B0D933 A0A0C9RP53 A0A0V0G777 V5HXY5 Q177G5 A0A1Q3FCB4 A0A1W4VVF8 Q29QH7 Q294B1 A0A1B2AKH8 B4QZ20 A0A3B0JSE3 A0A069DVF8 A0A0K8WD33 B4HDY7 A0A2R5LD34 A0A1B6FN20 A0A336LHM2 A0A2R5LD22 A0A1Z5L8S7 A0A1A9W720 A0A131XQF3 A0A1E1WYB7 A0A023G0W6 T1J3W1 A0A0P4XBK8 A0A0P5FXU6 A0A1S3CXS7 G3MTB5 G3MTF2 A0A023G9G4 A0A1A9UZ38 A0A1B6C1Q2 A0A087TZP7 H3AVT1 A0A1B6DCY2 A0A023FPB7 A0A1B0G090 A0A151IEN1 A0A195FBQ1 A0A3S1BZY7
A0A084VJB5 A0A182RNV2 A0A2P8YRS1 A0A182QXE5 A0A182N677 A0A067RPV2 A0A182PKP3 A0A182T987 A0A182Y470 D2A5N8 A0A1L8E127 A0A2M3ZKZ9 A0A1L8EBN4 A0A2M4C4A0 B3MX82 W5JHW8 A0A0K8TQ78 A0A182LYC1 A0A182JQ03 A0A182VHA3 A0A182TFI2 A0A182WRN7 Q7QD68 A0A182HU42 A0A182FRB4 A0A2M4AZC8 A0A023EEW8 A0A1I8PVB6 A0A1L8E9D5 A0A0T6B100 B4LZY6 N6U9S5 A0A0A9WSI4 A0A0P4VR50 R4FMS5 A0A0L0CPQ9 B4K5U7 B4NIS5 A0A1J1HX72 B4PS59 A0A0K8UJN7 B0W950 B3P0T8 A0A1I8M3C4 A0A293LFH9 R4WE56 A0A0K8R4F8 B7P5Y1 A0A2R7WBA6 A0A034WTF0 A0A1B0D933 A0A0C9RP53 A0A0V0G777 V5HXY5 Q177G5 A0A1Q3FCB4 A0A1W4VVF8 Q29QH7 Q294B1 A0A1B2AKH8 B4QZ20 A0A3B0JSE3 A0A069DVF8 A0A0K8WD33 B4HDY7 A0A2R5LD34 A0A1B6FN20 A0A336LHM2 A0A2R5LD22 A0A1Z5L8S7 A0A1A9W720 A0A131XQF3 A0A1E1WYB7 A0A023G0W6 T1J3W1 A0A0P4XBK8 A0A0P5FXU6 A0A1S3CXS7 G3MTB5 G3MTF2 A0A023G9G4 A0A1A9UZ38 A0A1B6C1Q2 A0A087TZP7 H3AVT1 A0A1B6DCY2 A0A023FPB7 A0A1B0G090 A0A151IEN1 A0A195FBQ1 A0A3S1BZY7
Ontologies
GO
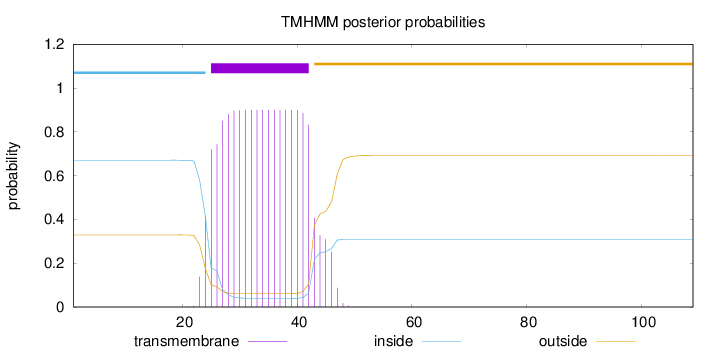
Topology
Length:
109
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
17.67601
Exp number, first 60 AAs:
17.67601
Total prob of N-in:
0.67069
POSSIBLE N-term signal
sequence
inside
1 - 24
TMhelix
25 - 42
outside
43 - 109
Population Genetic Test Statistics
Pi
133.533052
Theta
134.901731
Tajima's D
-0.519381
CLR
13.465833
CSRT
0.23938803059847
Interpretation
Uncertain
