Gene
KWMTBOMO12604
Pre Gene Modal
BGIBMGA001548
Annotation
PREDICTED:_tyrosine-protein_kinase_CSK_isoform_X1_[Bombyx_mori]
Full name
Tyrosine-protein kinase
+ More
Tyrosine-protein kinase CSK
Tyrosine-protein kinase CSK
Alternative Name
C-Src kinase
Protein-tyrosine kinase MPK-2
p50CSK
Protein-tyrosine kinase MPK-2
p50CSK
Location in the cell
Nuclear Reliability : 2.402
Sequence
CDS
ATGAATTCTGACGTGCATCGTCATCAGGCGCATCCGCCATTGGCACAGCACGGAGTGCACGGTGCGGCGCAGGGCGTGGGCCCTTGTGGCTGGTATTCGGCTTCTGCCGCTCCTGCACCCACGGCCCCTGCTCGTCTTAAGCGCGTACAAGCACCTCATCAGCTACCGTCTGGAGGGAACTTGAGCGGCAGTAATGGCCCGCACGCGCCTCTCTCCCCGTCCGCATTGCAGAATACTATACCGCACAATCTTATAATGTCATCGTCAGTCCGGACACCCGCTCCGGCCGCCACTGTACCGTCCGCCATGCACACCTCGCAAACAGCACCAGCAGTCACGAAAGGACAGAGCGATGTCTTGGTAAACTCGTCCGTACAGAACCCACCGCCGGCCGGCAACAGGCTGGTCAATATGACACAGTGGCCATGGCATCACGGCGCGATATCGAGGGAGCGAGCGGAGGCGTTGCTCTCCGGCTCTCCGGACGGGGTTTTCTTGGTGCGAGAGAGCACTAACTTCCCAGGAGACCACACGCTGTGCGTCCGATTCCGAGGAAGGGTGGAACACTATAGAGTAAAATGGGCGACAGCGCCCGACAACCAAAACCAACGTCTGACCATAGACGATGAAGAATTCTTCGATAACATGACGGACTTAATTCATCACTACCTGCAGGACGCGGACGGGCTGTGCACGAAGCTGGTGCGGTGCCTCCCCAAGGCGACGGCGAACGGCACCAACAACAACGGCGCCCCGCAGCAGCAGTACCCGCCCCACCCGCAGCAGCCGCCGCCGTATCCGCCCACGCCGAGCGTGCCGAGCGCGCTGGCGGCGGGACACTTCCCGCACGCGTACGTCGCGCCGGCCGCCTCCACCGACCAGACGGACGGCACCTTGCAGTCGCAAAGCTTCGTTGACGCGCGTTGGAAAATAGACGAGCGCGACCTTGAAATCCGAGAAAATATCGGCAAAGGAGAATTCGGCGACGTGATGCTAGGGATCCTGAACGGCACGCAGAAAGTGGCCGTCAAAATTCTCAAAGATCGGGAAGCCGCTAGTAAATTCCGCGCTGAAGCTAGTGTGATGGCATCGTTGAAGCACGAGAACCTGGTCCGTCTGCTGGGGCTGGTGTTCACGAGCAGCGGCGGCACGTGCCTGGTGACGGAGCACTGCGCGCAGGGCTCGCTGCTCGACTACCTGCGCAGCCGCGGCCGCCACTACGTCACGCAGCTCAACCAGATCAACTTCGCGTTTGACGCGTGCTGTGGGATGGAATATTTAGAAAGGCAACGTGTCGTACACCGCGACCTGGCGGCGAGAAATGTGCTCATCTCGGCGGAAGGCACCGCCAAGGTGGCGGACTTCGGTCTGGCGCGAGCTGGTGCAGCGCTGGAGGATCCGGCGGAGGCGGCTCGGCTACAGGCCAAGTTGCCCATTAAATGGACAGCACCGGAGGCACTTAAATATAACAAATTCTCAAACAAGTCGGATATGTGGAGTTTTGGAATTTTATTATGGGAAATTTACTCGTTTGGAAGAGTACCTTATCCTAGAATTCCGCTGGCGGAGGTGGTGCGGCACGTGGAGCGCGGCTACCGCATGGAGGCGCCGGAGGGGTGTCCGCCCGGCCCCTACGAGCTCATGCGCGCCGCCTGGCACGCCGACCCCGCCTCGCGACCCACCTTCCTGCACATGCGCCGCAACCTCGCCGACATACGCGACCACACGCCCGCGCAGGCTGATGATGATGATGATTCGCTGGTAGCCTAA
Protein
MNSDVHRHQAHPPLAQHGVHGAAQGVGPCGWYSASAAPAPTAPARLKRVQAPHQLPSGGNLSGSNGPHAPLSPSALQNTIPHNLIMSSSVRTPAPAATVPSAMHTSQTAPAVTKGQSDVLVNSSVQNPPPAGNRLVNMTQWPWHHGAISRERAEALLSGSPDGVFLVRESTNFPGDHTLCVRFRGRVEHYRVKWATAPDNQNQRLTIDDEEFFDNMTDLIHHYLQDADGLCTKLVRCLPKATANGTNNNGAPQQQYPPHPQQPPPYPPTPSVPSALAAGHFPHAYVAPAASTDQTDGTLQSQSFVDARWKIDERDLEIRENIGKGEFGDVMLGILNGTQKVAVKILKDREAASKFRAEASVMASLKHENLVRLLGLVFTSSGGTCLVTEHCAQGSLLDYLRSRGRHYVTQLNQINFAFDACCGMEYLERQRVVHRDLAARNVLISAEGTAKVADFGLARAGAALEDPAEAARLQAKLPIKWTAPEALKYNKFSNKSDMWSFGILLWEIYSFGRVPYPRIPLAEVVRHVERGYRMEAPEGCPPGPYELMRAAWHADPASRPTFLHMRRNLADIRDHTPAQADDDDDSLVA
Summary
Description
Non-receptor tyrosine-protein kinase that plays an important role in the regulation of cell growth, differentiation, migration and immune response. Phosphorylates tyrosine residues located in the C-terminal tails of Src-family kinases (SFKs) including LCK, SRC, HCK, FYN, LYN, CSK or YES1. Upon tail phosphorylation, Src-family members engage in intramolecular interactions between the phosphotyrosine tail and the SH2 domain that result in an inactive conformation. To inhibit SFKs, CSK is recruited to the plasma membrane via binding to transmembrane proteins or adapter proteins located near the plasma membrane. Suppresses signaling by various surface receptors, including T-cell receptor (TCR) and B-cell receptor (BCR) by phosphorylating and maintaining inactive several positive effectors such as FYN or LCK (By similarity).
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Subunit
Homodimer (via SH3-domain) (By similarity). Interacts with PTPN22 (PubMed:8890164). Interacts with phosphorylated SIT1, PAG1, LIME1 and TGFB1I1; these interactions serve to recruit CSK to the membrane where it can phosphorylate and inhibit Src-family kinases (PubMed:9858471, PubMed:12218089, PubMed:12612075, PubMed:16166631). Interacts with SRCIN1 (By similarity). Interacts with RHOH (By similarity). Interacts (via SH2 domain) with SCIMP (By similarity). Interacts (via SH2 domain) with PRAG1 (when phosphorylated at 'Tyr-391'); this interaction prevents translocation of CSK from the cytoplasm to the membrane leading to increased activity of CSK (By similarity).
Homodimer (via SH3-domain). Interacts with PTPN22. Interacts with phosphorylated SIT1, PAG1, LIME1 and TGFB1I1; these interactions serve to recruit CSK to the membrane where it can phosphorylate and inhibit Src-family kinases. Interacts with SRCIN1. Interacts with RHOH. Interacts (via SH2 domain) with SCIMP (By similarity). Interacts (via SH2 domain) with PRAG1 (when phosphorylated at 'Tyr-391'); this interaction prevents translocation of CSK from the cytoplasm to the membrane leading to increased activity of CSK (PubMed:21873224).
Homodimer (via SH3-domain). Interacts with PTPN22. Interacts with phosphorylated SIT1, PAG1, LIME1 and TGFB1I1; these interactions serve to recruit CSK to the membrane where it can phosphorylate and inhibit Src-family kinases. Interacts with SRCIN1. Interacts with RHOH. Interacts (via SH2 domain) with SCIMP (By similarity). Interacts (via SH2 domain) with PRAG1 (when phosphorylated at 'Tyr-391'); this interaction prevents translocation of CSK from the cytoplasm to the membrane leading to increased activity of CSK (PubMed:21873224).
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the protein kinase superfamily. Tyr protein kinase family. CSK subfamily.
Belongs to the protein kinase superfamily. Tyr protein kinase family. CSK subfamily.
Keywords
3D-structure
Acetylation
Adaptive immunity
ATP-binding
Cell membrane
Complete proteome
Cytoplasm
Direct protein sequencing
Immunity
Kinase
Magnesium
Manganese
Membrane
Metal-binding
Nucleotide-binding
Phosphoprotein
Reference proteome
SH2 domain
SH3 domain
Transferase
Tyrosine-protein kinase
Feature
chain Tyrosine-protein kinase CSK
Uniprot
H9IWB7
A0A2A4JS35
A0A194Q2D8
A0A212ERP4
A0A2H1VDH0
A0A2A4JQK7
+ More
A0A3S2M928 A0A2J7RIA3 A0A067QVQ7 A0A1B6JEY6 A0A1B6DC98 A0A1B6GL01 A0A023F4U1 A0A0V0GBU4 A0A224XRI6 A0A069DU21 A0A2R7WMY9 U5EGS0 B4JFH9 A0A088AI81 A0A2A3EBE0 A0A0A9YK91 A0A0L7QKJ6 A0A158N9Q5 A0A195F8C8 E0VB13 A0A195BNP3 E2BQA4 A0A151ISY3 F4WRI0 A0A151IC38 A0A151XGX3 A0A154PQC7 A0A1D2NB97 B4M474 A0A026WH98 E2AGI3 A0A0J7LBA3 E9J0V9 A0A0P6HX98 B3M100 A0A0P8Y024 A0A310S809 A0A2H8TCW2 J9KAY7 A0A1Y1LC76 A0A0N8DPS2 A0A076FKY1 L7M1U7 A0A224YUT6 A0A131YWK3 A0A131XCJ3 T1JDP9 A0A131XXI8 A0A087UEP1 A0A226F1H3 V5H7J5 A0A1E1XQP7 L7MKD7 A0A1E1X8H3 A0A2L2YK04 A0A2R5L9A0 A0A1Z5KWJ4 A0A443RF52 A0A443RF28 A0A232EXM9 A0A0P4VYG5 A0A1S3ICC4 A0A1V9X5D6 A0A443SQH9 Q3UVH2 A0A1D1V0P3 A0A1U7QKS8 M3VUK8 G3IIC0 A0A2U3Y9U0 A0A2Y9KEJ2 A0A3P4KYT3 M3Z0S9 G3WSI5 E2RFF7 A0A3Q7RDQ4 A0A3Q7RFS7 A0A340WB71 P41241 A0A2U3Y9U7 D2HKY4 A0A452FQZ5 A0A2Y9HIQ2 A0A340WGT5 A0A452SQW6 A0A3Q7TEX9 P32577 A0A2K6GVY3 A0A2Y9DHG3 F6QS08 A0A2U3WGL3
A0A3S2M928 A0A2J7RIA3 A0A067QVQ7 A0A1B6JEY6 A0A1B6DC98 A0A1B6GL01 A0A023F4U1 A0A0V0GBU4 A0A224XRI6 A0A069DU21 A0A2R7WMY9 U5EGS0 B4JFH9 A0A088AI81 A0A2A3EBE0 A0A0A9YK91 A0A0L7QKJ6 A0A158N9Q5 A0A195F8C8 E0VB13 A0A195BNP3 E2BQA4 A0A151ISY3 F4WRI0 A0A151IC38 A0A151XGX3 A0A154PQC7 A0A1D2NB97 B4M474 A0A026WH98 E2AGI3 A0A0J7LBA3 E9J0V9 A0A0P6HX98 B3M100 A0A0P8Y024 A0A310S809 A0A2H8TCW2 J9KAY7 A0A1Y1LC76 A0A0N8DPS2 A0A076FKY1 L7M1U7 A0A224YUT6 A0A131YWK3 A0A131XCJ3 T1JDP9 A0A131XXI8 A0A087UEP1 A0A226F1H3 V5H7J5 A0A1E1XQP7 L7MKD7 A0A1E1X8H3 A0A2L2YK04 A0A2R5L9A0 A0A1Z5KWJ4 A0A443RF52 A0A443RF28 A0A232EXM9 A0A0P4VYG5 A0A1S3ICC4 A0A1V9X5D6 A0A443SQH9 Q3UVH2 A0A1D1V0P3 A0A1U7QKS8 M3VUK8 G3IIC0 A0A2U3Y9U0 A0A2Y9KEJ2 A0A3P4KYT3 M3Z0S9 G3WSI5 E2RFF7 A0A3Q7RDQ4 A0A3Q7RFS7 A0A340WB71 P41241 A0A2U3Y9U7 D2HKY4 A0A452FQZ5 A0A2Y9HIQ2 A0A340WGT5 A0A452SQW6 A0A3Q7TEX9 P32577 A0A2K6GVY3 A0A2Y9DHG3 F6QS08 A0A2U3WGL3
EC Number
2.7.10.2
Pubmed
19121390
26354079
22118469
24845553
25474469
26334808
+ More
17994087 25401762 26823975 21347285 20566863 20798317 21719571 27289101 24508170 30249741 21282665 28004739 25576852 28797301 26830274 28049606 25765539 29209593 28503490 26561354 28528879 28648823 28327890 10349636 11042159 11076861 11217851 12466851 16141073 27649274 17975172 21804562 23236062 21709235 16341006 7511815 16141072 15489334 1281307 7685657 8890164 9858471 12218089 12612075 16166631 21183079 11685249 20010809 1709258 1722201 7515063 10801129 14613929 21873224 25243066
17994087 25401762 26823975 21347285 20566863 20798317 21719571 27289101 24508170 30249741 21282665 28004739 25576852 28797301 26830274 28049606 25765539 29209593 28503490 26561354 28528879 28648823 28327890 10349636 11042159 11076861 11217851 12466851 16141073 27649274 17975172 21804562 23236062 21709235 16341006 7511815 16141072 15489334 1281307 7685657 8890164 9858471 12218089 12612075 16166631 21183079 11685249 20010809 1709258 1722201 7515063 10801129 14613929 21873224 25243066
EMBL
BABH01006344
BABH01006345
NWSH01000780
PCG74283.1
KQ459564
KPI99706.1
+ More
AGBW02012992 OWR44121.1 ODYU01001967 SOQ38889.1 PCG74281.1 RSAL01000006 RVE54310.1 NEVH01003502 PNF40560.1 KK852898 KDR14137.1 GECU01031354 GECU01016690 GECU01010018 JAS76352.1 JAS91016.1 JAS97688.1 GEDC01026840 GEDC01013981 JAS10458.1 JAS23317.1 GECZ01029194 GECZ01016012 GECZ01006698 JAS40575.1 JAS53757.1 JAS63071.1 GBBI01002231 JAC16481.1 GECL01000561 JAP05563.1 GFTR01005805 JAW10621.1 GBGD01001697 JAC87192.1 KK855120 PTY20988.1 GANO01003329 JAB56542.1 CH916369 EDV93460.1 KZ288293 PBC29045.1 GBHO01042297 GBHO01042296 GBHO01042295 GBHO01037590 GBHO01037573 GBHO01027716 GBHO01027715 GBHO01027712 GBHO01027711 GBHO01025319 GBHO01025318 GBHO01025316 GBHO01025315 GBHO01013653 GBHO01013651 GBHO01013650 GBHO01013649 GBHO01013648 GBRD01008929 GBRD01008928 GDHC01013034 GDHC01007928 JAG01307.1 JAG01308.1 JAG01309.1 JAG06014.1 JAG06031.1 JAG15888.1 JAG15889.1 JAG15892.1 JAG15893.1 JAG18285.1 JAG18286.1 JAG18288.1 JAG18289.1 JAG29951.1 JAG29953.1 JAG29954.1 JAG29955.1 JAG29956.1 JAG56892.1 JAQ05595.1 JAQ10701.1 KQ414982 KOC59026.1 ADTU01009736 ADTU01009737 KQ981727 KYN36636.1 AAZO01000572 DS235020 EEB10569.1 KQ976428 KYM87842.1 GL449698 EFN82158.1 KQ981064 KYN09871.1 GL888287 EGI63197.1 KQ978067 KYM97655.1 KQ982169 KYQ59548.1 KQ435026 KZC13937.1 LJIJ01000108 ODN02521.1 CH940652 EDW59435.2 KK107211 QOIP01000005 EZA55338.1 RLU22694.1 GL439316 EFN67440.1 LBMM01000061 KMR05118.1 GL767538 EFZ13551.1 GDIP01187589 GDIQ01028461 LRGB01000642 JAJ35813.1 JAN66276.1 KZS17359.1 CH902617 EDV43229.2 KPU80147.1 KQ770716 OAD52628.1 GFXV01000151 MBW11956.1 ABLF02030156 GEZM01060095 GEZM01060094 JAV71269.1 GDIP01012871 JAM90844.1 KF516677 AII16581.1 GACK01006733 JAA58301.1 GFPF01006434 MAA17580.1 GEDV01005529 JAP83028.1 GEFH01004736 JAP63845.1 JH432107 GEFM01004861 JAP70935.1 KK119489 KFM75830.1 LNIX01000001 OXA62786.1 GANP01005373 JAB79095.1 GFAA01002163 JAU01272.1 GACK01000747 JAA64287.1 GFAC01003680 JAT95508.1 IAAA01036060 IAAA01036061 LAA08458.1 GGLE01001955 MBY06081.1 GFJQ02007511 JAV99458.1 NCKU01000860 RWS13873.1 NCKU01000861 RWS13870.1 NNAY01001730 OXU23103.1 GDRN01107837 JAI57365.1 MNPL01023305 OQR68809.1 NCKV01000784 RWS29753.1 AK137297 BAE23297.1 BDGG01000002 GAU93147.1 AANG04001761 JH002978 EGW15026.1 CYRY02001003 VCW55680.1 AEYP01046596 JP008159 AER96756.1 AEFK01001178 AEFK01001179 AEFK01001180 AAEX03016316 AAEX03016317 AAEX03016318 U05247 AY902339 AK156058 AK170815 BC018394 BC052006 X57242 ACTA01108031 ACTA01116031 GL192984 EFB20241.1 LWLT01000019 X58631 BC098863 GAMT01001202 GAMS01000600 GAMR01007519 GAMQ01006009 GAMP01004935 JAB10659.1 JAB22536.1 JAB26413.1 JAB35842.1 JAB47820.1
AGBW02012992 OWR44121.1 ODYU01001967 SOQ38889.1 PCG74281.1 RSAL01000006 RVE54310.1 NEVH01003502 PNF40560.1 KK852898 KDR14137.1 GECU01031354 GECU01016690 GECU01010018 JAS76352.1 JAS91016.1 JAS97688.1 GEDC01026840 GEDC01013981 JAS10458.1 JAS23317.1 GECZ01029194 GECZ01016012 GECZ01006698 JAS40575.1 JAS53757.1 JAS63071.1 GBBI01002231 JAC16481.1 GECL01000561 JAP05563.1 GFTR01005805 JAW10621.1 GBGD01001697 JAC87192.1 KK855120 PTY20988.1 GANO01003329 JAB56542.1 CH916369 EDV93460.1 KZ288293 PBC29045.1 GBHO01042297 GBHO01042296 GBHO01042295 GBHO01037590 GBHO01037573 GBHO01027716 GBHO01027715 GBHO01027712 GBHO01027711 GBHO01025319 GBHO01025318 GBHO01025316 GBHO01025315 GBHO01013653 GBHO01013651 GBHO01013650 GBHO01013649 GBHO01013648 GBRD01008929 GBRD01008928 GDHC01013034 GDHC01007928 JAG01307.1 JAG01308.1 JAG01309.1 JAG06014.1 JAG06031.1 JAG15888.1 JAG15889.1 JAG15892.1 JAG15893.1 JAG18285.1 JAG18286.1 JAG18288.1 JAG18289.1 JAG29951.1 JAG29953.1 JAG29954.1 JAG29955.1 JAG29956.1 JAG56892.1 JAQ05595.1 JAQ10701.1 KQ414982 KOC59026.1 ADTU01009736 ADTU01009737 KQ981727 KYN36636.1 AAZO01000572 DS235020 EEB10569.1 KQ976428 KYM87842.1 GL449698 EFN82158.1 KQ981064 KYN09871.1 GL888287 EGI63197.1 KQ978067 KYM97655.1 KQ982169 KYQ59548.1 KQ435026 KZC13937.1 LJIJ01000108 ODN02521.1 CH940652 EDW59435.2 KK107211 QOIP01000005 EZA55338.1 RLU22694.1 GL439316 EFN67440.1 LBMM01000061 KMR05118.1 GL767538 EFZ13551.1 GDIP01187589 GDIQ01028461 LRGB01000642 JAJ35813.1 JAN66276.1 KZS17359.1 CH902617 EDV43229.2 KPU80147.1 KQ770716 OAD52628.1 GFXV01000151 MBW11956.1 ABLF02030156 GEZM01060095 GEZM01060094 JAV71269.1 GDIP01012871 JAM90844.1 KF516677 AII16581.1 GACK01006733 JAA58301.1 GFPF01006434 MAA17580.1 GEDV01005529 JAP83028.1 GEFH01004736 JAP63845.1 JH432107 GEFM01004861 JAP70935.1 KK119489 KFM75830.1 LNIX01000001 OXA62786.1 GANP01005373 JAB79095.1 GFAA01002163 JAU01272.1 GACK01000747 JAA64287.1 GFAC01003680 JAT95508.1 IAAA01036060 IAAA01036061 LAA08458.1 GGLE01001955 MBY06081.1 GFJQ02007511 JAV99458.1 NCKU01000860 RWS13873.1 NCKU01000861 RWS13870.1 NNAY01001730 OXU23103.1 GDRN01107837 JAI57365.1 MNPL01023305 OQR68809.1 NCKV01000784 RWS29753.1 AK137297 BAE23297.1 BDGG01000002 GAU93147.1 AANG04001761 JH002978 EGW15026.1 CYRY02001003 VCW55680.1 AEYP01046596 JP008159 AER96756.1 AEFK01001178 AEFK01001179 AEFK01001180 AAEX03016316 AAEX03016317 AAEX03016318 U05247 AY902339 AK156058 AK170815 BC018394 BC052006 X57242 ACTA01108031 ACTA01116031 GL192984 EFB20241.1 LWLT01000019 X58631 BC098863 GAMT01001202 GAMS01000600 GAMR01007519 GAMQ01006009 GAMP01004935 JAB10659.1 JAB22536.1 JAB26413.1 JAB35842.1 JAB47820.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000283053
UP000235965
+ More
UP000027135 UP000001070 UP000005203 UP000242457 UP000053825 UP000005205 UP000078541 UP000009046 UP000078540 UP000008237 UP000078492 UP000007755 UP000078542 UP000075809 UP000076502 UP000094527 UP000008792 UP000053097 UP000279307 UP000000311 UP000036403 UP000076858 UP000007801 UP000007819 UP000054359 UP000198287 UP000285301 UP000215335 UP000085678 UP000192247 UP000288716 UP000186922 UP000189706 UP000011712 UP000001075 UP000245341 UP000248482 UP000000715 UP000007648 UP000002254 UP000286640 UP000286641 UP000265300 UP000000589 UP000008912 UP000291000 UP000248481 UP000291022 UP000286642 UP000002494 UP000233160 UP000248480 UP000008225 UP000245340
UP000027135 UP000001070 UP000005203 UP000242457 UP000053825 UP000005205 UP000078541 UP000009046 UP000078540 UP000008237 UP000078492 UP000007755 UP000078542 UP000075809 UP000076502 UP000094527 UP000008792 UP000053097 UP000279307 UP000000311 UP000036403 UP000076858 UP000007801 UP000007819 UP000054359 UP000198287 UP000285301 UP000215335 UP000085678 UP000192247 UP000288716 UP000186922 UP000189706 UP000011712 UP000001075 UP000245341 UP000248482 UP000000715 UP000007648 UP000002254 UP000286640 UP000286641 UP000265300 UP000000589 UP000008912 UP000291000 UP000248481 UP000291022 UP000286642 UP000002494 UP000233160 UP000248480 UP000008225 UP000245340
Interpro
IPR036860
SH2_dom_sf
+ More
IPR020635 Tyr_kinase_cat_dom
IPR008266 Tyr_kinase_AS
IPR000719 Prot_kinase_dom
IPR000980 SH2
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR017441 Protein_kinase_ATP_BS
IPR035027 Csk-like_SH2
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR026739 AP_beta
IPR016024 ARM-type_fold
IPR002553 Clathrin/coatomer_adapt-like_N
IPR029390 AP3B_C
IPR011989 ARM-like
IPR020635 Tyr_kinase_cat_dom
IPR008266 Tyr_kinase_AS
IPR000719 Prot_kinase_dom
IPR000980 SH2
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR017441 Protein_kinase_ATP_BS
IPR035027 Csk-like_SH2
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR026739 AP_beta
IPR016024 ARM-type_fold
IPR002553 Clathrin/coatomer_adapt-like_N
IPR029390 AP3B_C
IPR011989 ARM-like
Gene 3D
ProteinModelPortal
H9IWB7
A0A2A4JS35
A0A194Q2D8
A0A212ERP4
A0A2H1VDH0
A0A2A4JQK7
+ More
A0A3S2M928 A0A2J7RIA3 A0A067QVQ7 A0A1B6JEY6 A0A1B6DC98 A0A1B6GL01 A0A023F4U1 A0A0V0GBU4 A0A224XRI6 A0A069DU21 A0A2R7WMY9 U5EGS0 B4JFH9 A0A088AI81 A0A2A3EBE0 A0A0A9YK91 A0A0L7QKJ6 A0A158N9Q5 A0A195F8C8 E0VB13 A0A195BNP3 E2BQA4 A0A151ISY3 F4WRI0 A0A151IC38 A0A151XGX3 A0A154PQC7 A0A1D2NB97 B4M474 A0A026WH98 E2AGI3 A0A0J7LBA3 E9J0V9 A0A0P6HX98 B3M100 A0A0P8Y024 A0A310S809 A0A2H8TCW2 J9KAY7 A0A1Y1LC76 A0A0N8DPS2 A0A076FKY1 L7M1U7 A0A224YUT6 A0A131YWK3 A0A131XCJ3 T1JDP9 A0A131XXI8 A0A087UEP1 A0A226F1H3 V5H7J5 A0A1E1XQP7 L7MKD7 A0A1E1X8H3 A0A2L2YK04 A0A2R5L9A0 A0A1Z5KWJ4 A0A443RF52 A0A443RF28 A0A232EXM9 A0A0P4VYG5 A0A1S3ICC4 A0A1V9X5D6 A0A443SQH9 Q3UVH2 A0A1D1V0P3 A0A1U7QKS8 M3VUK8 G3IIC0 A0A2U3Y9U0 A0A2Y9KEJ2 A0A3P4KYT3 M3Z0S9 G3WSI5 E2RFF7 A0A3Q7RDQ4 A0A3Q7RFS7 A0A340WB71 P41241 A0A2U3Y9U7 D2HKY4 A0A452FQZ5 A0A2Y9HIQ2 A0A340WGT5 A0A452SQW6 A0A3Q7TEX9 P32577 A0A2K6GVY3 A0A2Y9DHG3 F6QS08 A0A2U3WGL3
A0A3S2M928 A0A2J7RIA3 A0A067QVQ7 A0A1B6JEY6 A0A1B6DC98 A0A1B6GL01 A0A023F4U1 A0A0V0GBU4 A0A224XRI6 A0A069DU21 A0A2R7WMY9 U5EGS0 B4JFH9 A0A088AI81 A0A2A3EBE0 A0A0A9YK91 A0A0L7QKJ6 A0A158N9Q5 A0A195F8C8 E0VB13 A0A195BNP3 E2BQA4 A0A151ISY3 F4WRI0 A0A151IC38 A0A151XGX3 A0A154PQC7 A0A1D2NB97 B4M474 A0A026WH98 E2AGI3 A0A0J7LBA3 E9J0V9 A0A0P6HX98 B3M100 A0A0P8Y024 A0A310S809 A0A2H8TCW2 J9KAY7 A0A1Y1LC76 A0A0N8DPS2 A0A076FKY1 L7M1U7 A0A224YUT6 A0A131YWK3 A0A131XCJ3 T1JDP9 A0A131XXI8 A0A087UEP1 A0A226F1H3 V5H7J5 A0A1E1XQP7 L7MKD7 A0A1E1X8H3 A0A2L2YK04 A0A2R5L9A0 A0A1Z5KWJ4 A0A443RF52 A0A443RF28 A0A232EXM9 A0A0P4VYG5 A0A1S3ICC4 A0A1V9X5D6 A0A443SQH9 Q3UVH2 A0A1D1V0P3 A0A1U7QKS8 M3VUK8 G3IIC0 A0A2U3Y9U0 A0A2Y9KEJ2 A0A3P4KYT3 M3Z0S9 G3WSI5 E2RFF7 A0A3Q7RDQ4 A0A3Q7RFS7 A0A340WB71 P41241 A0A2U3Y9U7 D2HKY4 A0A452FQZ5 A0A2Y9HIQ2 A0A340WGT5 A0A452SQW6 A0A3Q7TEX9 P32577 A0A2K6GVY3 A0A2Y9DHG3 F6QS08 A0A2U3WGL3
PDB
1K9A
E-value=5.91835e-112,
Score=1035
Ontologies
GO
GO:0005524
GO:0004715
GO:0045197
GO:0045571
GO:0048167
GO:0007293
GO:0008285
GO:0034334
GO:0035332
GO:0016477
GO:0043068
GO:0010669
GO:0016192
GO:0006886
GO:0030117
GO:0005886
GO:0042802
GO:0042997
GO:0034236
GO:0005911
GO:0034332
GO:0060368
GO:0010989
GO:0033673
GO:0045779
GO:0007420
GO:0046872
GO:0006468
GO:0070373
GO:0005737
GO:0050765
GO:0070064
GO:0046777
GO:0035556
GO:0004713
GO:0045121
GO:0016740
GO:0048709
GO:0050863
GO:0019903
GO:0071375
GO:0043406
GO:0032715
GO:0002250
GO:0004672
GO:0030286
GO:0008270
GO:0016787
GO:0004190
GO:0006508
GO:0016286
PANTHER
Topology
Subcellular location
Cytoplasm
Cell membrane
Cell membrane
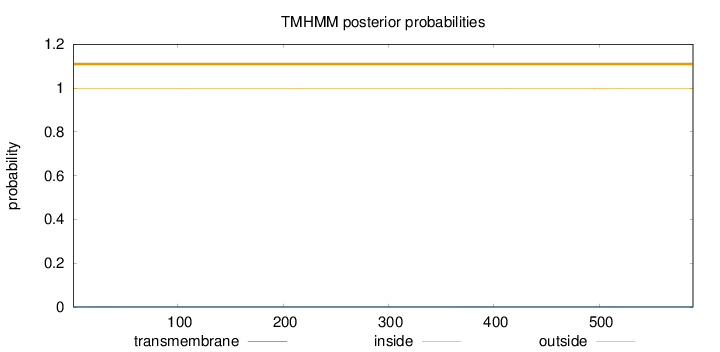
Length:
589
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03775
Exp number, first 60 AAs:
0.00796
Total prob of N-in:
0.00358
outside
1 - 589
Population Genetic Test Statistics
Pi
259.21564
Theta
173.741035
Tajima's D
1.254249
CLR
0.312771
CSRT
0.727963601819909
Interpretation
Uncertain
