Gene
KWMTBOMO12591
Annotation
PREDICTED:_uncharacterized_protein_LOC106119284?_partial_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 1.883
Sequence
CDS
ATGATTATGAGGTTGTGCGGCTGCGTCCCACATTTCTATAAAAAAATAGAAAACGAAAGAATATGCAGCCTACCTGAACTGAGATGCATTGTTCTTTACAAAAGAGAGATCACCACTCTAACAGCTTCAAACGAAACATTGCAAAGGTTTAACGATTATACCGATTTACCGAGAAGGTCACGCGACTGCGGATGTTTAGGTGATTGCGAAGCCGACGTCTACCAGCAAGATCCCGCGTCACTTTTGCCTCAAGAGTCATTGAACAGATTAAGGATCAGTGTCACGTCCTTTCCGAAAGTGAGATTTATGAGGGAAATTATGTTCAGTACTTATGACATTATACTACGGAGTGGCGGTATCGTGAACTTGTGTATCGGCACTTCGTTTATATCTATAATGGAACTGATGCTGACGGCCATAAGACTCCCAATCTACGAAATAGCGAACATAGCGCAGGCCTGTAGAAATGCTAAGACGGCGGATGCACAGAAAGTGAACAATAAAAAAATTAAATAG
Protein
MIMRLCGCVPHFYKKIENERICSLPELRCIVLYKREITTLTASNETLQRFNDYTDLPRRSRDCGCLGDCEADVYQQDPASLLPQESLNRLRISVTSFPKVRFMREIMFSTYDIILRSGGIVNLCIGTSFISIMELMLTAIRLPIYEIANIAQACRNAKTADAQKVNNKKIK
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
A0A194PZE7
N6U1R5
D7EHU3
A0A1Y1KQV7
A0A437B3V0
A0A1B6HI54
+ More
A0A0N0PBY3 A0A1Y1KRK2 H9ISX4 A0A2H1VKC3 A0A194PX82 A0A194R0M7 A0A182JQ12 A0A1B6D396 A0A0Q9W3Z0 A0A2R7X459 A0A2S2P0H9 B4KNP8 A0A182R4L2 A0A336L920 T1H7W7 A0A182PUD1 A0A0P9BZ20 A0A182UKQ3 A0A182YEH0 B4MS37 A0A084WEE5 A0A0B4LG42 A0A1W4ULV8 N6W4Z4 A0A0J9RJQ0 Q9W2B5 A0A1W4UZ35 A0A0M5J2D8 A0A1C7CZG4 A0A182WWV0 A0A2P8Z3P4 Q7PTD1 A0A3B0JPX1 A0A0T6BIH2 A0A0R1DXX9 A0A0Q5VZ50 A0A1I8NEQ8 A0A2P8ZFK0 A0A182WLE3 A0A1I8PIK6 A0A067QXR7 D6X1W4 J9K6J9 A0A1B0G173 A0A212F4Y2 A0A1A9V8K0 A0A182NJB3 A0A182M0L1 A0A1B0A345 A0A1I8N4R5 U4UF01 A0A336M002 K7J3E4 A0A1W4XL62 J9JJZ4 B4L2P1 A0A1W4XAZ8 A0A232F9M1 A0A1A9YDV1 K7INU5 A0A1W4XKA4 A0A1W4XAQ5 A0A310SQ88 A0A1B0C102 A0A232F8Q4 Q9VX46 B4Q2H6 B3NWV1 M9PHF4 F8QU26 K7JJP6 A0A0L0C8E2 A0A1A9W418 B4IF21 B4JKS3 A7URR0 A0A0M8ZQ73 B4MSE4 A0A1W4VVK3 A0A2J7Q831 A0A0N0U3N5 A0A0L7L8G0 A0A2S2R5A1 A0A067QUS6 B4M1E1 A0A2J7PQL0 U5EW41 A0A0L7RDT9 B0XDP8 A0A0M4EKC4 A0A182GXU4 A0A1B0AAZ8
A0A0N0PBY3 A0A1Y1KRK2 H9ISX4 A0A2H1VKC3 A0A194PX82 A0A194R0M7 A0A182JQ12 A0A1B6D396 A0A0Q9W3Z0 A0A2R7X459 A0A2S2P0H9 B4KNP8 A0A182R4L2 A0A336L920 T1H7W7 A0A182PUD1 A0A0P9BZ20 A0A182UKQ3 A0A182YEH0 B4MS37 A0A084WEE5 A0A0B4LG42 A0A1W4ULV8 N6W4Z4 A0A0J9RJQ0 Q9W2B5 A0A1W4UZ35 A0A0M5J2D8 A0A1C7CZG4 A0A182WWV0 A0A2P8Z3P4 Q7PTD1 A0A3B0JPX1 A0A0T6BIH2 A0A0R1DXX9 A0A0Q5VZ50 A0A1I8NEQ8 A0A2P8ZFK0 A0A182WLE3 A0A1I8PIK6 A0A067QXR7 D6X1W4 J9K6J9 A0A1B0G173 A0A212F4Y2 A0A1A9V8K0 A0A182NJB3 A0A182M0L1 A0A1B0A345 A0A1I8N4R5 U4UF01 A0A336M002 K7J3E4 A0A1W4XL62 J9JJZ4 B4L2P1 A0A1W4XAZ8 A0A232F9M1 A0A1A9YDV1 K7INU5 A0A1W4XKA4 A0A1W4XAQ5 A0A310SQ88 A0A1B0C102 A0A232F8Q4 Q9VX46 B4Q2H6 B3NWV1 M9PHF4 F8QU26 K7JJP6 A0A0L0C8E2 A0A1A9W418 B4IF21 B4JKS3 A7URR0 A0A0M8ZQ73 B4MSE4 A0A1W4VVK3 A0A2J7Q831 A0A0N0U3N5 A0A0L7L8G0 A0A2S2R5A1 A0A067QUS6 B4M1E1 A0A2J7PQL0 U5EW41 A0A0L7RDT9 B0XDP8 A0A0M4EKC4 A0A182GXU4 A0A1B0AAZ8
Pubmed
26354079
23537049
18362917
19820115
28004739
19121390
+ More
17994087 25244985 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 22936249 20966253 29403074 12364791 14747013 17210077 17550304 25315136 24845553 22118469 20075255 28648823 26108605 26227816 26483478
17994087 25244985 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 22936249 20966253 29403074 12364791 14747013 17210077 17550304 25315136 24845553 22118469 20075255 28648823 26108605 26227816 26483478
EMBL
KQ459585
KPI98418.1
APGK01043502
KB741014
ENN75495.1
KQ973114
+ More
EFA12114.2 GEZM01080733 JAV62065.1 RSAL01000175 RVE45049.1 GECU01033329 JAS74377.1 KQ460864 KPJ11815.1 GEZM01080732 JAV62066.1 BABH01013035 ODYU01003023 SOQ41265.1 KQ459586 KPI97932.1 KQ460883 KPJ11348.1 GEDC01017185 JAS20113.1 CH940648 KRF79677.1 KK856919 PTY26579.1 GGMR01010350 MBY22969.1 CH933808 EDW10033.2 UFQS01002201 UFQT01002201 SSX13481.1 SSX32911.1 ACPB03000915 CH902619 KPU76597.1 CH963850 EDW74926.2 ATLV01023206 KE525341 KFB48589.1 AE013599 AHN56495.1 CM000071 ENO01847.2 CM002911 KMY95679.1 AAF46777.2 CP012524 ALC40968.1 PYGN01000211 PSN51115.1 AAAB01008807 EAA04078.5 OUUW01000001 SPP75396.1 LJIG01000245 KRT86695.1 CM000158 KRJ99686.1 CH954179 KQS62314.1 PYGN01000073 PSN55272.1 KK852851 KDR15054.1 KQ971371 EFA10172.2 ABLF02031579 ABLF02031581 CCAG010016192 AGBW02010291 OWR48797.1 AXCM01003059 KB632293 ERL91617.1 UFQT01000343 SSX23390.1 ABLF02037336 CH933810 EDW07839.1 NNAY01000660 OXU27138.1 KQ761346 OAD57838.1 JXJN01023841 JXJN01023842 NNAY01000645 OXU27221.1 AE014298 AAF48734.2 CM000162 EDX02617.2 CH954180 EDV46498.2 AGB95488.1 HM026485 ADL60138.1 JRES01000866 KNC27674.1 CH480832 EDW46275.1 CH916370 EDW00176.1 EDO64702.1 KQ435922 KOX68687.1 CH963851 EDW75033.2 NEVH01016978 PNF24738.1 KQ435881 KOX69844.1 JTDY01002246 KOB71808.1 GGMS01015972 MBY85175.1 KK852925 KDR13715.1 CH940651 EDW65495.2 NEVH01022635 PNF18623.1 GANO01003129 JAB56742.1 KQ414612 KOC69152.1 DS232771 EDS45556.1 CP012528 ALC49816.1 JXUM01096276 KQ564253 KXJ72523.1
EFA12114.2 GEZM01080733 JAV62065.1 RSAL01000175 RVE45049.1 GECU01033329 JAS74377.1 KQ460864 KPJ11815.1 GEZM01080732 JAV62066.1 BABH01013035 ODYU01003023 SOQ41265.1 KQ459586 KPI97932.1 KQ460883 KPJ11348.1 GEDC01017185 JAS20113.1 CH940648 KRF79677.1 KK856919 PTY26579.1 GGMR01010350 MBY22969.1 CH933808 EDW10033.2 UFQS01002201 UFQT01002201 SSX13481.1 SSX32911.1 ACPB03000915 CH902619 KPU76597.1 CH963850 EDW74926.2 ATLV01023206 KE525341 KFB48589.1 AE013599 AHN56495.1 CM000071 ENO01847.2 CM002911 KMY95679.1 AAF46777.2 CP012524 ALC40968.1 PYGN01000211 PSN51115.1 AAAB01008807 EAA04078.5 OUUW01000001 SPP75396.1 LJIG01000245 KRT86695.1 CM000158 KRJ99686.1 CH954179 KQS62314.1 PYGN01000073 PSN55272.1 KK852851 KDR15054.1 KQ971371 EFA10172.2 ABLF02031579 ABLF02031581 CCAG010016192 AGBW02010291 OWR48797.1 AXCM01003059 KB632293 ERL91617.1 UFQT01000343 SSX23390.1 ABLF02037336 CH933810 EDW07839.1 NNAY01000660 OXU27138.1 KQ761346 OAD57838.1 JXJN01023841 JXJN01023842 NNAY01000645 OXU27221.1 AE014298 AAF48734.2 CM000162 EDX02617.2 CH954180 EDV46498.2 AGB95488.1 HM026485 ADL60138.1 JRES01000866 KNC27674.1 CH480832 EDW46275.1 CH916370 EDW00176.1 EDO64702.1 KQ435922 KOX68687.1 CH963851 EDW75033.2 NEVH01016978 PNF24738.1 KQ435881 KOX69844.1 JTDY01002246 KOB71808.1 GGMS01015972 MBY85175.1 KK852925 KDR13715.1 CH940651 EDW65495.2 NEVH01022635 PNF18623.1 GANO01003129 JAB56742.1 KQ414612 KOC69152.1 DS232771 EDS45556.1 CP012528 ALC49816.1 JXUM01096276 KQ564253 KXJ72523.1
Proteomes
UP000053268
UP000019118
UP000007266
UP000283053
UP000053240
UP000005204
+ More
UP000075881 UP000008792 UP000009192 UP000075900 UP000015103 UP000075885 UP000007801 UP000075902 UP000076408 UP000007798 UP000030765 UP000000803 UP000192221 UP000001819 UP000092553 UP000075882 UP000076407 UP000245037 UP000007062 UP000268350 UP000002282 UP000008711 UP000095301 UP000075920 UP000095300 UP000027135 UP000007819 UP000092444 UP000007151 UP000078200 UP000075884 UP000075883 UP000092445 UP000030742 UP000002358 UP000192223 UP000215335 UP000092443 UP000092460 UP000037069 UP000091820 UP000001292 UP000001070 UP000053105 UP000235965 UP000037510 UP000053825 UP000002320 UP000069940 UP000249989
UP000075881 UP000008792 UP000009192 UP000075900 UP000015103 UP000075885 UP000007801 UP000075902 UP000076408 UP000007798 UP000030765 UP000000803 UP000192221 UP000001819 UP000092553 UP000075882 UP000076407 UP000245037 UP000007062 UP000268350 UP000002282 UP000008711 UP000095301 UP000075920 UP000095300 UP000027135 UP000007819 UP000092444 UP000007151 UP000078200 UP000075884 UP000075883 UP000092445 UP000030742 UP000002358 UP000192223 UP000215335 UP000092443 UP000092460 UP000037069 UP000091820 UP000001292 UP000001070 UP000053105 UP000235965 UP000037510 UP000053825 UP000002320 UP000069940 UP000249989
Pfam
PF00858 ASC
ProteinModelPortal
A0A194PZE7
N6U1R5
D7EHU3
A0A1Y1KQV7
A0A437B3V0
A0A1B6HI54
+ More
A0A0N0PBY3 A0A1Y1KRK2 H9ISX4 A0A2H1VKC3 A0A194PX82 A0A194R0M7 A0A182JQ12 A0A1B6D396 A0A0Q9W3Z0 A0A2R7X459 A0A2S2P0H9 B4KNP8 A0A182R4L2 A0A336L920 T1H7W7 A0A182PUD1 A0A0P9BZ20 A0A182UKQ3 A0A182YEH0 B4MS37 A0A084WEE5 A0A0B4LG42 A0A1W4ULV8 N6W4Z4 A0A0J9RJQ0 Q9W2B5 A0A1W4UZ35 A0A0M5J2D8 A0A1C7CZG4 A0A182WWV0 A0A2P8Z3P4 Q7PTD1 A0A3B0JPX1 A0A0T6BIH2 A0A0R1DXX9 A0A0Q5VZ50 A0A1I8NEQ8 A0A2P8ZFK0 A0A182WLE3 A0A1I8PIK6 A0A067QXR7 D6X1W4 J9K6J9 A0A1B0G173 A0A212F4Y2 A0A1A9V8K0 A0A182NJB3 A0A182M0L1 A0A1B0A345 A0A1I8N4R5 U4UF01 A0A336M002 K7J3E4 A0A1W4XL62 J9JJZ4 B4L2P1 A0A1W4XAZ8 A0A232F9M1 A0A1A9YDV1 K7INU5 A0A1W4XKA4 A0A1W4XAQ5 A0A310SQ88 A0A1B0C102 A0A232F8Q4 Q9VX46 B4Q2H6 B3NWV1 M9PHF4 F8QU26 K7JJP6 A0A0L0C8E2 A0A1A9W418 B4IF21 B4JKS3 A7URR0 A0A0M8ZQ73 B4MSE4 A0A1W4VVK3 A0A2J7Q831 A0A0N0U3N5 A0A0L7L8G0 A0A2S2R5A1 A0A067QUS6 B4M1E1 A0A2J7PQL0 U5EW41 A0A0L7RDT9 B0XDP8 A0A0M4EKC4 A0A182GXU4 A0A1B0AAZ8
A0A0N0PBY3 A0A1Y1KRK2 H9ISX4 A0A2H1VKC3 A0A194PX82 A0A194R0M7 A0A182JQ12 A0A1B6D396 A0A0Q9W3Z0 A0A2R7X459 A0A2S2P0H9 B4KNP8 A0A182R4L2 A0A336L920 T1H7W7 A0A182PUD1 A0A0P9BZ20 A0A182UKQ3 A0A182YEH0 B4MS37 A0A084WEE5 A0A0B4LG42 A0A1W4ULV8 N6W4Z4 A0A0J9RJQ0 Q9W2B5 A0A1W4UZ35 A0A0M5J2D8 A0A1C7CZG4 A0A182WWV0 A0A2P8Z3P4 Q7PTD1 A0A3B0JPX1 A0A0T6BIH2 A0A0R1DXX9 A0A0Q5VZ50 A0A1I8NEQ8 A0A2P8ZFK0 A0A182WLE3 A0A1I8PIK6 A0A067QXR7 D6X1W4 J9K6J9 A0A1B0G173 A0A212F4Y2 A0A1A9V8K0 A0A182NJB3 A0A182M0L1 A0A1B0A345 A0A1I8N4R5 U4UF01 A0A336M002 K7J3E4 A0A1W4XL62 J9JJZ4 B4L2P1 A0A1W4XAZ8 A0A232F9M1 A0A1A9YDV1 K7INU5 A0A1W4XKA4 A0A1W4XAQ5 A0A310SQ88 A0A1B0C102 A0A232F8Q4 Q9VX46 B4Q2H6 B3NWV1 M9PHF4 F8QU26 K7JJP6 A0A0L0C8E2 A0A1A9W418 B4IF21 B4JKS3 A7URR0 A0A0M8ZQ73 B4MSE4 A0A1W4VVK3 A0A2J7Q831 A0A0N0U3N5 A0A0L7L8G0 A0A2S2R5A1 A0A067QUS6 B4M1E1 A0A2J7PQL0 U5EW41 A0A0L7RDT9 B0XDP8 A0A0M4EKC4 A0A182GXU4 A0A1B0AAZ8
Ontologies
GO
PANTHER
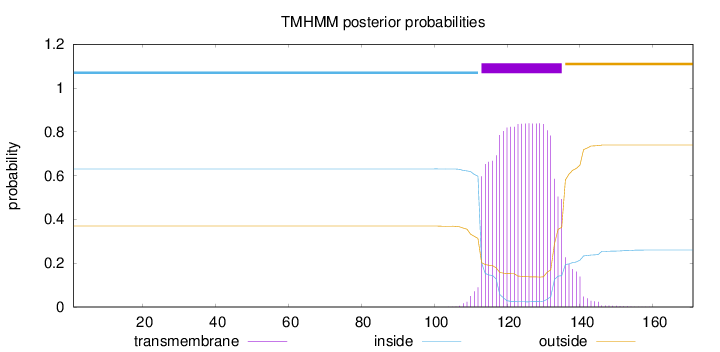
Topology
Length:
171
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.55684
Exp number, first 60 AAs:
0
Total prob of N-in:
0.63063
inside
1 - 112
TMhelix
113 - 135
outside
136 - 171
Population Genetic Test Statistics
Pi
47.854746
Theta
118.131297
Tajima's D
-1.960991
CLR
188.37758
CSRT
0.0175991200439978
Interpretation
Uncertain
