Pre Gene Modal
BGIBMGA001430
Annotation
hypothetical_protein_KGM_21105_[Danaus_plexippus]
Full name
Integrator complex subunit 3 homolog
+ More
Integrator complex subunit 3
Integrator complex subunit 3
Alternative Name
SOSS complex subunit A homolog
SOSS complex subunit A
Sensor of single-strand DNA complex subunit A
SOSS complex subunit A
Sensor of single-strand DNA complex subunit A
Location in the cell
Nuclear Reliability : 3.419
Sequence
CDS
ATGGAGCAGGCCAAATCTAATTCACCCAAACTATTTGTGAGCACCGCAATAGAAAATAAGGATGAAGTGGAAGAGAGGTACGAAAGGGCTTACAACTATTTTCAATCTCTTGTAGCGGACTGCACCGAAAAAGAAGCACATGACGCCCTTAATAACGCAGTCTGTAAAAATCATGAGGAAGTTTCAGTTGGCATGCTCATGTCTATACTTACAGAACCGCAGAATGCTACAAAATGTTACAGAGATCTTACACTTATAACCAGAGATGGACTAACGTGTGTGCTTAATAATCTATCCAATTTAATTTTAGAAAGGTACTTAAAGTTTTCGGACACTACAAGGAACCAATTAATGTGGCTAATAAAAGAGATGATTAGAAATGCAGTTACAGGAATAGATAATGTTTGTTGGAATTTAATGCGATACGCATCCGGAGGCGATATAACACCCAAAAATATTATATTAGTAGAATCTCTGTTGGACATGTTTATTGAACATAGAGGTTGGTTGGATAAGTTCCCTGTACTTATATGCATGGTTGTGTATACATACTTAAGGCTTGTAGAAGACCATAATATACCCCAGTTAATGAACCTAAGACAGAAAGAAGTTAACTTTGTTATCACTCTGATAAGGGAACGTTTTACTGATGTCTTATCTTTGGGAAGAGACTTAGTAAGACTGCTTCAAAATGTTGCTCGCATACCAGAGTTCAATCAGCTTTGGCAGGATATCTTGGTCAATCCCAAATCATTAAGCCCTACATTTACCAATGTGCTACAATTGTTACAAACACGAACATCTAGAAGGTATCTTCAGTCAAGATTAACTCCGGACATGGAAAGGAAATTAGTTTTTCTAACTTCACAAGTTCGGTTCGGTCACCACAAAAAGTATCAAGAATGGTTCCAGAGACAATATCTTGCCACACCAGAGTCACAATCACTAAGAAGTGATATGATAAGGTTTATTGTTGGAGTAATTCATCCCACTAATGAACTTCTCTGTTCTGATATAATTCCCAGATGGGCAGTGATTGGTTGGCTCCTTACAACTTGTACGTCAAATGTAGCTGCATCAAATGCTAAATTGGCCCTGTTTTATGATTGGCTTTTTTATGACCCTGAAAAAGACAACATTATGAATATAGAACCTGCCATCCTTGTTATGCACCATTCAATGAGATCACATCCAGCAGTGACTGCAACATTATTAGATTTCTTGTGTCGAATCATTCCAAACTTCTACCCACCATATGCAGAGAAAGTTAGACAAGGAATATTCAATTCCCTTCAACAAATAATTGAGAAAAGGGTTTTGCCAAATTTGCATCCTCTGTTTGACTCTCAAAAGTTGGACCGAGAATTGCGTTCCACTGTAAGAGAAACCTTTAAAGAATTTTGTACGAATGGGAATGGTGAAGGTGTATCTGGCATACGCGATGAGGGTACTGATGATGCAATACCAAGAGTTGGGTCCGATGAACCAGCATTCTCGGATGATGAAGAAGAGACCATTACACCAACTATAGCAGAAGATACTGATGATGATGATTTACCTTTATCAGAAGTTCGTGCCAGAGAAAGACCTGAACTAGCTGCTGCACTGCCGTTAGTTTTGCGACCACTAGCAGAAACGCTTCTGGATGAGCGAACATCATCTTCCGCAACTGCTTTGCTTGATGCATGCGGATCTCCATCGCCAACGCCGAAAGCTCTCGCAGATCTTTTTGCTGCTGCGGTTCAAGAAGCAACACCTCTACGTATAAGCAACAATCCCGCTCCAGCCGAATTAGATAATGCAATAAATTCACCGCTTTTTTCCATGTTCAATTATCTAATAGACTCGAGTAGTGGAGCTGAACATAAACGGAAACTGGTATTAGAAACGTTCAAAGAAATCCACAGTCAAGGTTCTCTGAGATGTGTCGGTTTTAGCCTACTGTTTTATTTAAGAGTCTGCTACGAAAGAGATCGTCGTTCAGAACGAGATTCGGCGAAAAGACGTAACGTGAAATTCAAAGCTGACGTTTATAAGGACTACTGTAGTTATTTAGAATTAAAATTAGCCGAGAGCTTAGCGGACGATTTCGAAAGGTGCCAAGAATACGACACCAATGTTTTAGTTTGGTTAATACCAGATGTTTATAGAGAGTTCAAAGACCAAGCACAGAATCATATAAGATTATTGCATGCAATCGTATCGACCCTAGATGCGTGTCAACTTCAAAGACTAATCTGTTTGACATTGCAAGGAAATTTAATGATGTTCAAATCGGACGATATCACGACAATGTTGTCGACCAGCCTTGGATGGGAGACGTTCGAACAGTATTGTCTGTGGCAACTGTTAACCGCCCATGACATATCCGTTGAGGATGTTTTACCGATCATTCCGAAGTTATCATTCAAAGAGCATTCGGAGGCGCTCACGTCCGTTCTGTTGATGTTGAAGCAGGAAAGACCGACCGCCGATGTCGTAAGACAGTTGTTCTGTAGAACATACGACGAGGAGGATACATCGGTGGTTTCAACGTTGATGTATTGGTGCCAAGATTACGAAGATAAAGTCGGTGATTTAGTCGCGGCATTGTTGAGTACGCGCTATCCGGGTACTAGTCCGAACAAGCGAAAACGTCCAGGGAAACACAGCATGCCACCAAACGCACCGCCGTCTGCAGAGTTGGTGCTCAAACATTTGGAGCAGATGAGAATGTGCTGCGTTGAACAAGACGACATGTCGATTTTCAACATGGAATGTATGCAGAAGGCCCTACAACAAGCTCAATCGAATAGTAGCGATAGTTTAAGGGAAGAAGAGATTGTGAAGCCGAAACAAGCTAAACGTCGCAAGAAGGCAATTTCGGACAGTGATTGA
Protein
MEQAKSNSPKLFVSTAIENKDEVEERYERAYNYFQSLVADCTEKEAHDALNNAVCKNHEEVSVGMLMSILTEPQNATKCYRDLTLITRDGLTCVLNNLSNLILERYLKFSDTTRNQLMWLIKEMIRNAVTGIDNVCWNLMRYASGGDITPKNIILVESLLDMFIEHRGWLDKFPVLICMVVYTYLRLVEDHNIPQLMNLRQKEVNFVITLIRERFTDVLSLGRDLVRLLQNVARIPEFNQLWQDILVNPKSLSPTFTNVLQLLQTRTSRRYLQSRLTPDMERKLVFLTSQVRFGHHKKYQEWFQRQYLATPESQSLRSDMIRFIVGVIHPTNELLCSDIIPRWAVIGWLLTTCTSNVAASNAKLALFYDWLFYDPEKDNIMNIEPAILVMHHSMRSHPAVTATLLDFLCRIIPNFYPPYAEKVRQGIFNSLQQIIEKRVLPNLHPLFDSQKLDRELRSTVRETFKEFCTNGNGEGVSGIRDEGTDDAIPRVGSDEPAFSDDEEETITPTIAEDTDDDDLPLSEVRARERPELAAALPLVLRPLAETLLDERTSSSATALLDACGSPSPTPKALADLFAAAVQEATPLRISNNPAPAELDNAINSPLFSMFNYLIDSSSGAEHKRKLVLETFKEIHSQGSLRCVGFSLLFYLRVCYERDRRSERDSAKRRNVKFKADVYKDYCSYLELKLAESLADDFERCQEYDTNVLVWLIPDVYREFKDQAQNHIRLLHAIVSTLDACQLQRLICLTLQGNLMMFKSDDITTMLSTSLGWETFEQYCLWQLLTAHDISVEDVLPIIPKLSFKEHSEALTSVLLMLKQERPTADVVRQLFCRTYDEEDTSVVSTLMYWCQDYEDKVGDLVAALLSTRYPGTSPNKRKRPGKHSMPPNAPPSAELVLKHLEQMRMCCVEQDDMSIFNMECMQKALQQAQSNSSDSLREEEIVKPKQAKRRKKAISDSD
Summary
Description
Component of the Integrator (INT) complex. The Integrator complex is involved in the small nuclear RNAs (snRNA) U1 and U2 transcription and in their 3'-box-dependent processing. The Integrator complex is associated with the C-terminal domain (CTD) of RNA polymerase II largest subunit (POLR2A) and is recruited to the U1 and U2 snRNAs genes. Mediates recruitment of cytoplasmic dynein to the nuclear envelope, probably as component of the INT complex.
Component of the SOSS complex, a multiprotein complex that functions downstream of the MRN complex to promote DNA repair and G2/M checkpoint. The SOSS complex associates with single-stranded DNA at DNA lesions and influences diverse endpoints in the cellular DNA damage response including cell-cycle checkpoint activation, recombinational repair and maintenance of genomic stability. The SOSS complex is required for efficient homologous recombination-dependent repair of double-strand breaks (DSBs) and ATM-dependent signaling pathways. In the SOSS complex, it is required for the assembly of the complex and for stabilization of the complex at DNA damage sites.
Component of the Integrator (INT) complex. The Integrator complex is involved in the small nuclear RNAs (snRNA) U1 and U2 transcription and in their 3'-box-dependent processing. The Integrator complex is associated with the C-terminal domain (CTD) of RNA polymerase II largest subunit (POLR2A) and is recruited to the U1 and U2 snRNAs genes (Probable). Mediates recruitment of cytoplasmic dynein to the nuclear envelope, probably as component of the INT complex (PubMed:23904267).
Component of the SOSS complex, a multiprotein complex that functions downstream of the MRN complex to promote DNA repair and G2/M checkpoint. The SOSS complex associates with single-stranded DNA at DNA lesions and influences diverse endpoints in the cellular DNA damage response including cell-cycle checkpoint activation, recombinational repair and maintenance of genomic stability. The SOSS complex is required for efficient homologous recombination-dependent repair of double-strand breaks (DSBs) and ATM-dependent signaling pathways. In the SOSS complex, it is required for the assembly of the complex and for stabilization of the complex at DNA damage sites.
Component of the Integrator (INT) complex. The Integrator complex is involved in the small nuclear RNAs (snRNA) U1 and U2 transcription and in their 3'-box-dependent processing. The Integrator complex is associated with the C-terminal domain (CTD) of RNA polymerase II largest subunit (POLR2A) and is recruited to the U1 and U2 snRNAs genes (Probable). Mediates recruitment of cytoplasmic dynein to the nuclear envelope, probably as component of the INT complex (PubMed:23904267).
Subunit
Belongs to the multiprotein complex Integrator, at least composed of INTS1, INTS2, INTS3, INTS4, INTS5, INTS6, INTS7, INTS8, INTS9/RC74, INTS10, INTS11/CPSF3L and INTS12. Component of the SOSS complex, composed of SOSS-B (SOSS-B1/NABP2 or SOSS-B2/NABP1), SOSS-A/INTS3 and SOSS-C/INIP. SOSS complexes containing SOSS-B1/NABP2 are more abundant than complexes containing SOSS-B2/NABP1. Interacts with SOSS-B1/NABP2, SOSS-B2/NABP1 and SOSS-C/INIP; the interaction is direct. Interacts with NBN/NBS1.
Similarity
Belongs to the Integrator subunit 3 family.
Keywords
Complete proteome
Reference proteome
Acetylation
Alternative splicing
Cytoplasm
DNA damage
DNA repair
Nucleus
Phosphoprotein
3D-structure
Feature
chain Integrator complex subunit 3 homolog
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2A4K4U2
A0A437BVL4
A0A212F3U1
A0A1E1WFN2
A0A2H1VQF5
A0A0N0PCE9
+ More
H9IVZ9 A0A194PYK7 A0A0L7KRA7 E9IJD0 A0A0T6AVJ4 A0A154PE96 A0A0C9RWS3 A0A1Y1K7X9 D7EIT4 T1JE28 A0A336LPS8 A0A2R5LCI0 K1PWS0 A0A293MSH0 A0A443SMN7 Q7PRB8 A0A1S4FIC1 Q16ZT1 A0A2S2NCM0 E0VJF0 Q7TPD0 A0A0G2JFJ6 D3ZUT9 H0WHU8 A0A1U7R3H9 A0A2K5F4C3 A0A2I2ULH2 Q68E01-2 G3RTH1 A0A2R9B031 A0A2K6TQ05 F7FD65 H2Q040 G1SNY3 A0A2K5UIL5 A0A2K6A6F9 A0A2K6PKE6 A0A2K5NWS7 A0A2K5NWU9 A0A2J8VFQ4 H9FYM8 I3LX47 K9INJ5 G3SQX3 A0A2R9B497 A0A2I3SIQ7 A0A1U7TWR7 A0A384CK60 A0A2I2Y401 G1M097 A0A2K6A6I1 A0A2Y9E8S7 A0A452S0D1 A0A3Q7UWT3 A0A384CJ68 A0A151PIU9 G3HC14 F1SFW4 A0A2K5HVX4 S7MVJ6 A0A1S3F256 A0A2K5HVY7 Q5RE70 A0A1A6HYF6 A0A3Q7N0W0 A0A2Y9IH09 A0A2U3W4T5 G7MDS3 Q68E01 G7NUH7 A0A3Q7SC41 A0A452G4J0 A0A2Y9FVJ9 A0A2Y9GIS2 W5NSM6 L9JEB6 A0A383ZUL2 F6U224 E1BN70 A0A341BM90 A0A2Y9M790 A0A340XFU3 A0A0P6JU00 F6VZW0 A0A2K6PKC5 A0A1U7TBU4 A0A091E0W9 A0A452GFB4 A0A452GFB7 S9YGK6
H9IVZ9 A0A194PYK7 A0A0L7KRA7 E9IJD0 A0A0T6AVJ4 A0A154PE96 A0A0C9RWS3 A0A1Y1K7X9 D7EIT4 T1JE28 A0A336LPS8 A0A2R5LCI0 K1PWS0 A0A293MSH0 A0A443SMN7 Q7PRB8 A0A1S4FIC1 Q16ZT1 A0A2S2NCM0 E0VJF0 Q7TPD0 A0A0G2JFJ6 D3ZUT9 H0WHU8 A0A1U7R3H9 A0A2K5F4C3 A0A2I2ULH2 Q68E01-2 G3RTH1 A0A2R9B031 A0A2K6TQ05 F7FD65 H2Q040 G1SNY3 A0A2K5UIL5 A0A2K6A6F9 A0A2K6PKE6 A0A2K5NWS7 A0A2K5NWU9 A0A2J8VFQ4 H9FYM8 I3LX47 K9INJ5 G3SQX3 A0A2R9B497 A0A2I3SIQ7 A0A1U7TWR7 A0A384CK60 A0A2I2Y401 G1M097 A0A2K6A6I1 A0A2Y9E8S7 A0A452S0D1 A0A3Q7UWT3 A0A384CJ68 A0A151PIU9 G3HC14 F1SFW4 A0A2K5HVX4 S7MVJ6 A0A1S3F256 A0A2K5HVY7 Q5RE70 A0A1A6HYF6 A0A3Q7N0W0 A0A2Y9IH09 A0A2U3W4T5 G7MDS3 Q68E01 G7NUH7 A0A3Q7SC41 A0A452G4J0 A0A2Y9FVJ9 A0A2Y9GIS2 W5NSM6 L9JEB6 A0A383ZUL2 F6U224 E1BN70 A0A341BM90 A0A2Y9M790 A0A340XFU3 A0A0P6JU00 F6VZW0 A0A2K6PKC5 A0A1U7TBU4 A0A091E0W9 A0A452GFB4 A0A452GFB7 S9YGK6
Pubmed
22118469
26354079
19121390
26227816
21282665
28004739
+ More
18362917 19820115 22992520 12364791 17510324 20566863 16141072 15489334 21183079 19468303 15057822 22673903 17975172 17974005 14702039 16710414 16239144 18669648 19413330 19605351 19683501 20068231 21269460 21406692 22814378 23186163 23904267 24275569 22398555 22722832 25243066 16136131 21993624 25362486 17431167 25319552 20010809 24813606 22293439 21804562 30723633 22002653 20809919 23385571 19892987 19393038 17495919 28562605 23149746
18362917 19820115 22992520 12364791 17510324 20566863 16141072 15489334 21183079 19468303 15057822 22673903 17975172 17974005 14702039 16710414 16239144 18669648 19413330 19605351 19683501 20068231 21269460 21406692 22814378 23186163 23904267 24275569 22398555 22722832 25243066 16136131 21993624 25362486 17431167 25319552 20010809 24813606 22293439 21804562 30723633 22002653 20809919 23385571 19892987 19393038 17495919 28562605 23149746
EMBL
NWSH01000145
PCG79089.1
RSAL01000006
RVE54301.1
AGBW02010495
OWR48394.1
+ More
GDQN01005244 JAT85810.1 ODYU01003826 SOQ43075.1 KQ460622 KPJ13515.1 BABH01006289 KQ459585 KPI98416.1 JTDY01006988 KOB65549.1 GL763764 EFZ19350.1 LJIG01022765 KRT78830.1 KQ434886 KZC10189.1 GBYB01013510 JAG83277.1 GEZM01096470 JAV54877.1 KQ971380 EFA12368.2 JH432114 UFQT01000108 SSX20104.1 GGLE01003062 MBY07188.1 JH816741 EKC20815.1 GFWV01019056 MAA43784.1 NCKV01001215 RWS28789.1 AAAB01008859 CH477484 EAT40190.1 GGMR01002258 MBY14877.1 DS235222 EEB13506.1 AK040147 AK043206 BC003209 BC055344 AC145082 AABR07072757 AAQR03179064 AAQR03179065 AAQR03179066 AAQR03179067 AAQR03179068 AAQR03179069 AAQR03179070 AAQR03179071 AAQR03179072 AAQR03179073 AANG04002876 AL832133 AK290025 AK298746 AK304874 BX640786 BX640950 CR627233 CR749212 CR749376 AL513523 CH471121 BC025254 BC054513 BC073985 BC098431 BC105092 BC105094 BC116458 AK025572 AK074979 BK005722 CABD030006536 CABD030006537 AJFE02058273 AJFE02058274 AJFE02058275 AJFE02058276 AJFE02058277 AJFE02058278 AJFE02058279 AJFE02058280 AJFE02058281 GAMT01000778 GAMS01004005 GAMR01006515 GAMQ01005464 GAMP01002380 JAB11083.1 JAB19131.1 JAB27417.1 JAB36387.1 JAB50375.1 AC194541 GABC01002017 GABF01007250 GABD01008896 GABE01003229 NBAG03000084 JAA09321.1 JAA14895.1 JAA24204.1 JAA41510.1 PNI90820.1 PNI90824.1 AAGW02000507 AAGW02000508 AQIA01002370 AQIA01002371 ABGA01216453 ABGA01216454 ABGA01216455 ABGA01216456 ABGA01216457 ABGA01216458 ABGA01216459 ABGA01216460 ABGA01216461 NDHI03003420 PNJ56340.1 PNJ56341.1 JSUE03002457 JSUE03002458 JSUE03002459 JSUE03002460 JU335984 JU476866 JV047974 AFE79737.1 AFH33670.1 AFI38045.1 AGTP01066446 AGTP01066447 GABZ01003985 JAA49540.1 ACTA01073328 ACTA01081328 ACTA01089328 AKHW03000146 KYO48980.1 JH000275 EGV98136.1 AEMK02000022 DQIR01098998 HDA54474.1 KE162497 EPQ08536.1 CR857667 LZPO01007946 OBS83276.1 CM001253 EHH15270.1 CM001276 EHH50315.1 LWLT01000004 AMGL01004773 KB321128 ELW47322.1 GEBF01006224 JAN97408.1 KN121497 KFO36343.1 KB016598 EPY86576.1
GDQN01005244 JAT85810.1 ODYU01003826 SOQ43075.1 KQ460622 KPJ13515.1 BABH01006289 KQ459585 KPI98416.1 JTDY01006988 KOB65549.1 GL763764 EFZ19350.1 LJIG01022765 KRT78830.1 KQ434886 KZC10189.1 GBYB01013510 JAG83277.1 GEZM01096470 JAV54877.1 KQ971380 EFA12368.2 JH432114 UFQT01000108 SSX20104.1 GGLE01003062 MBY07188.1 JH816741 EKC20815.1 GFWV01019056 MAA43784.1 NCKV01001215 RWS28789.1 AAAB01008859 CH477484 EAT40190.1 GGMR01002258 MBY14877.1 DS235222 EEB13506.1 AK040147 AK043206 BC003209 BC055344 AC145082 AABR07072757 AAQR03179064 AAQR03179065 AAQR03179066 AAQR03179067 AAQR03179068 AAQR03179069 AAQR03179070 AAQR03179071 AAQR03179072 AAQR03179073 AANG04002876 AL832133 AK290025 AK298746 AK304874 BX640786 BX640950 CR627233 CR749212 CR749376 AL513523 CH471121 BC025254 BC054513 BC073985 BC098431 BC105092 BC105094 BC116458 AK025572 AK074979 BK005722 CABD030006536 CABD030006537 AJFE02058273 AJFE02058274 AJFE02058275 AJFE02058276 AJFE02058277 AJFE02058278 AJFE02058279 AJFE02058280 AJFE02058281 GAMT01000778 GAMS01004005 GAMR01006515 GAMQ01005464 GAMP01002380 JAB11083.1 JAB19131.1 JAB27417.1 JAB36387.1 JAB50375.1 AC194541 GABC01002017 GABF01007250 GABD01008896 GABE01003229 NBAG03000084 JAA09321.1 JAA14895.1 JAA24204.1 JAA41510.1 PNI90820.1 PNI90824.1 AAGW02000507 AAGW02000508 AQIA01002370 AQIA01002371 ABGA01216453 ABGA01216454 ABGA01216455 ABGA01216456 ABGA01216457 ABGA01216458 ABGA01216459 ABGA01216460 ABGA01216461 NDHI03003420 PNJ56340.1 PNJ56341.1 JSUE03002457 JSUE03002458 JSUE03002459 JSUE03002460 JU335984 JU476866 JV047974 AFE79737.1 AFH33670.1 AFI38045.1 AGTP01066446 AGTP01066447 GABZ01003985 JAA49540.1 ACTA01073328 ACTA01081328 ACTA01089328 AKHW03000146 KYO48980.1 JH000275 EGV98136.1 AEMK02000022 DQIR01098998 HDA54474.1 KE162497 EPQ08536.1 CR857667 LZPO01007946 OBS83276.1 CM001253 EHH15270.1 CM001276 EHH50315.1 LWLT01000004 AMGL01004773 KB321128 ELW47322.1 GEBF01006224 JAN97408.1 KN121497 KFO36343.1 KB016598 EPY86576.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053240
UP000005204
UP000053268
+ More
UP000037510 UP000076502 UP000007266 UP000005408 UP000288716 UP000007062 UP000008820 UP000009046 UP000000589 UP000002494 UP000005225 UP000189706 UP000233020 UP000011712 UP000005640 UP000001519 UP000240080 UP000233220 UP000008225 UP000002277 UP000001811 UP000233100 UP000233140 UP000233200 UP000233060 UP000001595 UP000006718 UP000005215 UP000007646 UP000189704 UP000261680 UP000008912 UP000248480 UP000291022 UP000286642 UP000291021 UP000050525 UP000001075 UP000008227 UP000233080 UP000081671 UP000092124 UP000286641 UP000248482 UP000245340 UP000009130 UP000286640 UP000291000 UP000248484 UP000248481 UP000002356 UP000011518 UP000261681 UP000002281 UP000009136 UP000252040 UP000248483 UP000265300 UP000002280 UP000028990 UP000291020
UP000037510 UP000076502 UP000007266 UP000005408 UP000288716 UP000007062 UP000008820 UP000009046 UP000000589 UP000002494 UP000005225 UP000189706 UP000233020 UP000011712 UP000005640 UP000001519 UP000240080 UP000233220 UP000008225 UP000002277 UP000001811 UP000233100 UP000233140 UP000233200 UP000233060 UP000001595 UP000006718 UP000005215 UP000007646 UP000189704 UP000261680 UP000008912 UP000248480 UP000291022 UP000286642 UP000291021 UP000050525 UP000001075 UP000008227 UP000233080 UP000081671 UP000092124 UP000286641 UP000248482 UP000245340 UP000009130 UP000286640 UP000291000 UP000248484 UP000248481 UP000002356 UP000011518 UP000261681 UP000002281 UP000009136 UP000252040 UP000248483 UP000265300 UP000002280 UP000028990 UP000291020
Interpro
IPR016024
ARM-type_fold
+ More
IPR019333 Int_cplx_su3
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR036116 FN3_sf
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR042099 AMP-dep_Synthh-like_sf
IPR000873 AMP-dep_Synth/Lig
IPR020845 AMP-binding_CS
IPR025110 AMP-bd_C
IPR019333 Int_cplx_su3
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR036116 FN3_sf
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR042099 AMP-dep_Synthh-like_sf
IPR000873 AMP-dep_Synth/Lig
IPR020845 AMP-binding_CS
IPR025110 AMP-bd_C
Gene 3D
CDD
ProteinModelPortal
A0A2A4K4U2
A0A437BVL4
A0A212F3U1
A0A1E1WFN2
A0A2H1VQF5
A0A0N0PCE9
+ More
H9IVZ9 A0A194PYK7 A0A0L7KRA7 E9IJD0 A0A0T6AVJ4 A0A154PE96 A0A0C9RWS3 A0A1Y1K7X9 D7EIT4 T1JE28 A0A336LPS8 A0A2R5LCI0 K1PWS0 A0A293MSH0 A0A443SMN7 Q7PRB8 A0A1S4FIC1 Q16ZT1 A0A2S2NCM0 E0VJF0 Q7TPD0 A0A0G2JFJ6 D3ZUT9 H0WHU8 A0A1U7R3H9 A0A2K5F4C3 A0A2I2ULH2 Q68E01-2 G3RTH1 A0A2R9B031 A0A2K6TQ05 F7FD65 H2Q040 G1SNY3 A0A2K5UIL5 A0A2K6A6F9 A0A2K6PKE6 A0A2K5NWS7 A0A2K5NWU9 A0A2J8VFQ4 H9FYM8 I3LX47 K9INJ5 G3SQX3 A0A2R9B497 A0A2I3SIQ7 A0A1U7TWR7 A0A384CK60 A0A2I2Y401 G1M097 A0A2K6A6I1 A0A2Y9E8S7 A0A452S0D1 A0A3Q7UWT3 A0A384CJ68 A0A151PIU9 G3HC14 F1SFW4 A0A2K5HVX4 S7MVJ6 A0A1S3F256 A0A2K5HVY7 Q5RE70 A0A1A6HYF6 A0A3Q7N0W0 A0A2Y9IH09 A0A2U3W4T5 G7MDS3 Q68E01 G7NUH7 A0A3Q7SC41 A0A452G4J0 A0A2Y9FVJ9 A0A2Y9GIS2 W5NSM6 L9JEB6 A0A383ZUL2 F6U224 E1BN70 A0A341BM90 A0A2Y9M790 A0A340XFU3 A0A0P6JU00 F6VZW0 A0A2K6PKC5 A0A1U7TBU4 A0A091E0W9 A0A452GFB4 A0A452GFB7 S9YGK6
H9IVZ9 A0A194PYK7 A0A0L7KRA7 E9IJD0 A0A0T6AVJ4 A0A154PE96 A0A0C9RWS3 A0A1Y1K7X9 D7EIT4 T1JE28 A0A336LPS8 A0A2R5LCI0 K1PWS0 A0A293MSH0 A0A443SMN7 Q7PRB8 A0A1S4FIC1 Q16ZT1 A0A2S2NCM0 E0VJF0 Q7TPD0 A0A0G2JFJ6 D3ZUT9 H0WHU8 A0A1U7R3H9 A0A2K5F4C3 A0A2I2ULH2 Q68E01-2 G3RTH1 A0A2R9B031 A0A2K6TQ05 F7FD65 H2Q040 G1SNY3 A0A2K5UIL5 A0A2K6A6F9 A0A2K6PKE6 A0A2K5NWS7 A0A2K5NWU9 A0A2J8VFQ4 H9FYM8 I3LX47 K9INJ5 G3SQX3 A0A2R9B497 A0A2I3SIQ7 A0A1U7TWR7 A0A384CK60 A0A2I2Y401 G1M097 A0A2K6A6I1 A0A2Y9E8S7 A0A452S0D1 A0A3Q7UWT3 A0A384CJ68 A0A151PIU9 G3HC14 F1SFW4 A0A2K5HVX4 S7MVJ6 A0A1S3F256 A0A2K5HVY7 Q5RE70 A0A1A6HYF6 A0A3Q7N0W0 A0A2Y9IH09 A0A2U3W4T5 G7MDS3 Q68E01 G7NUH7 A0A3Q7SC41 A0A452G4J0 A0A2Y9FVJ9 A0A2Y9GIS2 W5NSM6 L9JEB6 A0A383ZUL2 F6U224 E1BN70 A0A341BM90 A0A2Y9M790 A0A340XFU3 A0A0P6JU00 F6VZW0 A0A2K6PKC5 A0A1U7TBU4 A0A091E0W9 A0A452GFB4 A0A452GFB7 S9YGK6
PDB
4OWT
E-value=0,
Score=1631
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Localizes to nuclear foci following DNA damage. With evidence from 2 publications.
Cytoplasm Localizes to nuclear foci following DNA damage. With evidence from 2 publications.
Cytoplasm Localizes to nuclear foci following DNA damage. With evidence from 2 publications.
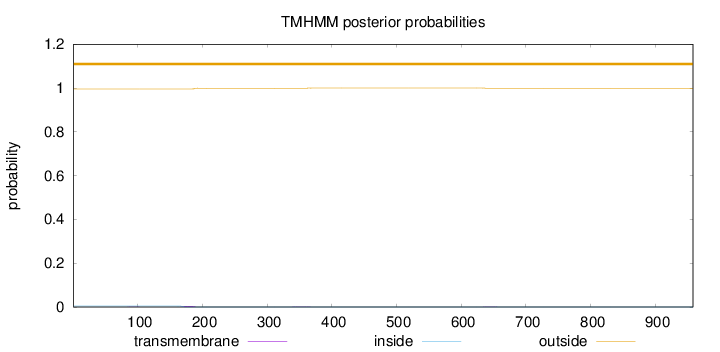
Length:
958
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10008
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00450
outside
1 - 958
Population Genetic Test Statistics
Pi
184.102007
Theta
176.790333
Tajima's D
0.412542
CLR
1.805278
CSRT
0.488725563721814
Interpretation
Uncertain
