Gene
KWMTBOMO12585
Pre Gene Modal
BGIBMGA001435
Annotation
PREDICTED:_A_disintegrin_and_metalloproteinase_with_thrombospondin_motifs_7_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 1.859 Nuclear Reliability : 2.418
Sequence
CDS
ATGCGCGCGAGTGAATCTCGGGTGCTCAACAAGATGGAAGTGCGGTGTTCATTGTTCAAACTCTTGCTTTTCCAAATGCTGGTAGCGTGGTGCGACGCGAAGCACGTCTGGACTGATGACATGACGAGAGTGGAGCTTGGTTCCGACGACGTTAACCGAGAAGTCCAGGATAGCATAGATACATTTATGCACACTGGTATCTACTCCCATCGACACCTGGATACGTCGCAGGTGCAAGTGGTGCGCCCGTTGAAAGTGACCCGTGATGGTGAGCTGGTCTCCCACGTGGTGGATCACTCCCACGAACACGGGCACGCGAGGGCGCGCCGTGACCTGCATCACACGGAGCATCTGCAGCCGCACTCTCTTCACTATAACCTCACCGTTGACGGCAGACAAATCAGGCTGGACCTAAGGCCGTCGGTGACATTCATTACTCCCGCAATGGTAGTGGAGCGACACAGCGAGCGCGGGCGCACGCGAGAACAGCCGAGGATCGAAGCCACGGCCTGCCACTACACGGGCTCTGTACGCGGACAGCCTGCATCCAGCGTAGCTCTTTCTGCTTGTGATGGACTTGCGGGCTTATTAAGGACAGAACAGGGTGAATATTGGATTGAACCTTCTACCCAAAAACCGTCTGATAATTCAGAAGGAAGACCACACGTTATTTTCAAACGATCAGCAGTAGACAAGGTTGCAGCATATCATAGAGCAAAAAGGGCTGTAGATAAAAATACCAATACCAACAGCAAAGTTAATCATGATAAAGACACCCAGAGATACACCAGCCAACACGCGAATCTCAGAAATTACCGTGAAAGTAAAGAAGAGAGAGACAGACAACGTAGGGAATATTTAGAACAGAGACAAAAAAGACTAGAGGCTATGAGGCGTAACCCTGTCGAATATAGAAGACAACAATCAAATTTACGAATGGAAGAAAGAAGAATGCACACGTATCCGAAAAGCAATTCGATTGAAGGTAGCAGATCATTGCATTCAAATCAGGGCCGTAAAAGAAATCAAAACAAAAGTGAAAGGCGATTGCGACGTAAACACCGAAGAAGAAGGAAACGTGCCGCGAAAAACTGTGGTACACGACAGCCGCCTTATCAATGGAAGCAAAAAAAACTTCACACTTCCGAGTTAGACGACAACAAGAAATACCGCTATAATAAGAACAACCGGTATAGCCGCCATCATCCAACCCGCTACAATTTAGACCCAAACAGGCGAGCGCCTCGTTCCGTGAGCAAGCCGCGGCACGTTGAGGTTTTACTCGTCGCAGACAAATCCATGTCGGAGTTCCACGACCAAGACAGCTTGGAAATTTACTTGTTGACTATAATGAATATGGTATCTTCTCTGTATATGGATCCGTCGATTGGAAACTACATTAAAGTGGTCGTTGTAAAAATTATCCTTGTCGAAGAGTTAGCTGCCGCACAGGAGCTAGAAGTGTCTACGAATGCTGATTCGACACTCGCATCCTTCTGCCGTTGGCAAGAGCAATTGAACCCAGACAACGATACAAACCCGCATCACCATGATGTCGCTATTTTAGTTACAAGACGCGACATCTGCAGCCAGCACGATTCTCCTTGTAGTACTTTGGGTGTAGCACACGTAGCGGGTATGTGCAAGCCGGACCGCAGCTGTTCTGTGAACGAAGACAACGGAATAATGCTCGCTCACACAATTACACACGAGCTGGGACACAATTTCGGTCTTTATCACGACACCGAAAAGATAGGCTGCCAAAGACGCGTGGGCTCCACACTGCACATCATGACACCTATATTTGAAGCAGACACCGTACAAGTAGCCTGGTCGCGTTGTAGTAAGCGAGACGTCACCAATTTCTTGGACGCCGGTTTGGGTGAATGTTTGAGCGACAGACCATCACAAGACGAATACGTGTATCCAGAAGTACCAGCTGGCGTAATGTTTGATGCGGCTTCACAATGTCACCTGCAGTTCGGCGCGGAAGCCGGCCTATGTTCTAAGCTGTCAGAGTTATGCGAACACCTATGGTGTCTGGTTGACAATGTGTGTACTACTATGATGCGTCCTGCAGCACCTGGCACCACTTGCGGACCCGATATGTGGTGTCAGAATCAGACCTGTGTGGCTCGGACTCCTTCACCGGCGCCTCGAGACGGCGGTTGGGGTCCATGGAGCGAGTGGAGTGAATGTTCTAGAACTTGCGGCGCTGGAGTATCCACCCAGTCGAGGGAATGCAATAACCCGGAGCCAGTCAATAATGGAAACTATTGTATTGGAGATCGTAGCAGGTACAAAGTGTGCAACACGGATCCGTGCCCTATCAACGAACCGTCATTCAGGGAAGTTCAGTGCTCCAGGCACAATAACATGCCATATAAGAATGAAACTATAGATGAATGGGTTCCCTACTTTGATCAAGATAAACCTTGCGAGTTACAATGCGTGCCGCGCGATGGTACTGATGTGGAGATGTTGGGAGGATTCGTCGCAGACGGTACTCCTTGCAGGCAAGCGATCGGGTCGCGGGACATGTGCATTTCTGGTGTGTGCTACAAAGTAGGTTGCGATTGGATTGTTGATTCTGACGTTGAAGAAGACGCATGCGGCGTCTGTGGGGGCGACGGAACAGCATGTAAAACTGTGCAAGGGATTTACACGAAAGGCACAACTAGACAATCAGGATTCAGTGAGGTTGCAATTATACCGGCAGGTTCGAGGAACGTGAAAATACAAGAGAAAGTTAGCCCTGGAAATTATATTTCGATCGGAAGTACAAAATCGAGAAGAATGTTTCTTAATGGAGCTCGAAACGCCACGTTGACGGAGTACTTCGTGGCTGGAGCTCAAGCTATTTACGAACGAGACCGTGACTGGGAAAAAGTTCGAATATCGGGACCGCTCGCCGAAGATATAAAAGTTTATCAACGTATATTCCGAGGACGACATCGCAACCCCGGCGTCACATACCAGTACACGGTCGACAAACAAAGACTTCACAAGAAACACAGCTACAGACTGTCCGAATGGACGCCGTGCTCCGTCACCTGCGGAGTTGGATACACGAGAAGATATTACGAATGCATTGATCAACATCAACGAGTGGTAGAACAATCCCAGTGCTACCACATCGAGCCGCCGCGACACCAGGCCCTCATGAAGCAGTGCAGGAAGCCGGCGTGCGTCCACTGGTGGGTCGGCGCGTGGAACCCTTGCTCTAAGCTTTGCCACATGCCGGGCGAGGAAGTCACTAAAGAACGCAGCGTGTACTGCGTCGATAAAGTAATGAACAAAGTAGTCGATGACAGCGAATGCGGCCAAGTGATTAAACCAATAACGACCATTAAATGTGCGGACGTGCCTGCGTGCTAA
Protein
MRASESRVLNKMEVRCSLFKLLLFQMLVAWCDAKHVWTDDMTRVELGSDDVNREVQDSIDTFMHTGIYSHRHLDTSQVQVVRPLKVTRDGELVSHVVDHSHEHGHARARRDLHHTEHLQPHSLHYNLTVDGRQIRLDLRPSVTFITPAMVVERHSERGRTREQPRIEATACHYTGSVRGQPASSVALSACDGLAGLLRTEQGEYWIEPSTQKPSDNSEGRPHVIFKRSAVDKVAAYHRAKRAVDKNTNTNSKVNHDKDTQRYTSQHANLRNYRESKEERDRQRREYLEQRQKRLEAMRRNPVEYRRQQSNLRMEERRMHTYPKSNSIEGSRSLHSNQGRKRNQNKSERRLRRKHRRRRKRAAKNCGTRQPPYQWKQKKLHTSELDDNKKYRYNKNNRYSRHHPTRYNLDPNRRAPRSVSKPRHVEVLLVADKSMSEFHDQDSLEIYLLTIMNMVSSLYMDPSIGNYIKVVVVKIILVEELAAAQELEVSTNADSTLASFCRWQEQLNPDNDTNPHHHDVAILVTRRDICSQHDSPCSTLGVAHVAGMCKPDRSCSVNEDNGIMLAHTITHELGHNFGLYHDTEKIGCQRRVGSTLHIMTPIFEADTVQVAWSRCSKRDVTNFLDAGLGECLSDRPSQDEYVYPEVPAGVMFDAASQCHLQFGAEAGLCSKLSELCEHLWCLVDNVCTTMMRPAAPGTTCGPDMWCQNQTCVARTPSPAPRDGGWGPWSEWSECSRTCGAGVSTQSRECNNPEPVNNGNYCIGDRSRYKVCNTDPCPINEPSFREVQCSRHNNMPYKNETIDEWVPYFDQDKPCELQCVPRDGTDVEMLGGFVADGTPCRQAIGSRDMCISGVCYKVGCDWIVDSDVEEDACGVCGGDGTACKTVQGIYTKGTTRQSGFSEVAIIPAGSRNVKIQEKVSPGNYISIGSTKSRRMFLNGARNATLTEYFVAGAQAIYERDRDWEKVRISGPLAEDIKVYQRIFRGRHRNPGVTYQYTVDKQRLHKKHSYRLSEWTPCSVTCGVGYTRRYYECIDQHQRVVEQSQCYHIEPPRHQALMKQCRKPACVHWWVGAWNPCSKLCHMPGEEVTKERSVYCVDKVMNKVVDDSECGQVIKPITTIKCADVPAC
Summary
Uniprot
EMBL
Proteomes
Pfam
Interpro
SUPFAM
SSF82895
SSF82895
Gene 3D
ProteinModelPortal
PDB
2RJQ
E-value=1.39636e-71,
Score=690
Ontologies
KEGG
GO
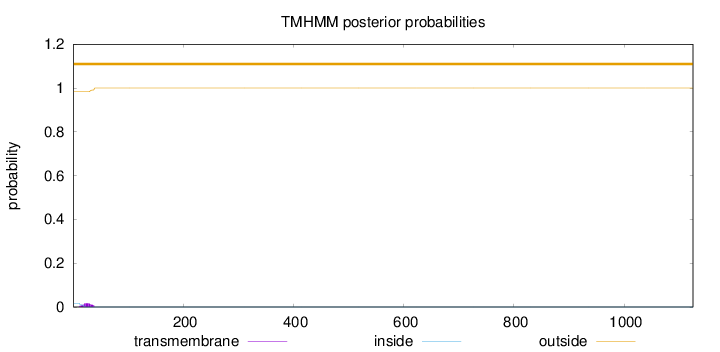
Topology
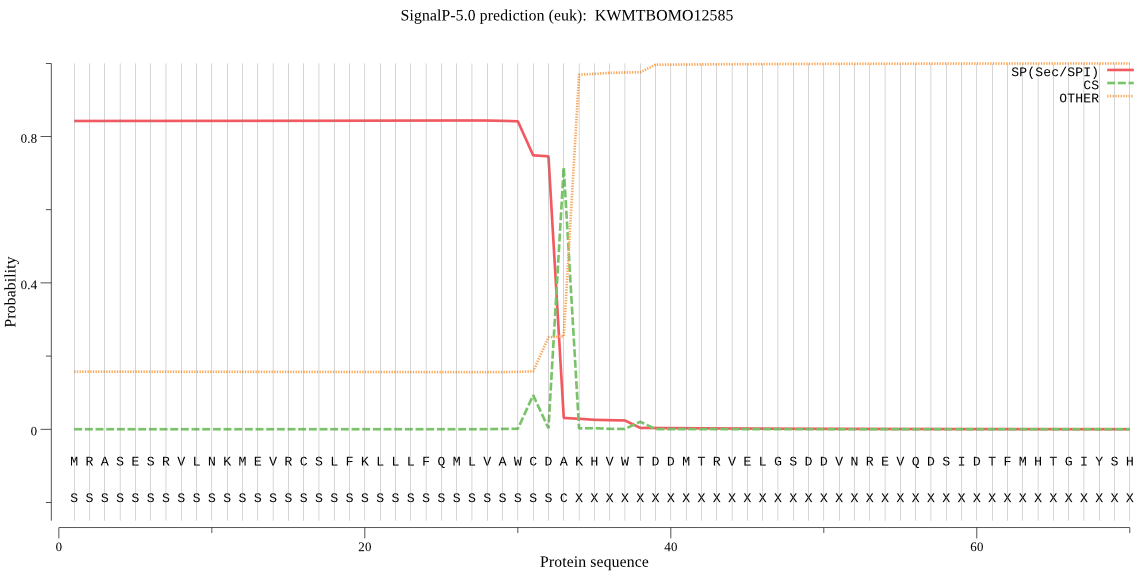
SignalP
Position: 1 - 33,
Likelihood: 0.843732
Length:
1125
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.32854
Exp number, first 60 AAs:
0.32592
Total prob of N-in:
0.01723
outside
1 - 1125
Population Genetic Test Statistics
Pi
184.953278
Theta
166.142201
Tajima's D
0.329285
CLR
0.808764
CSRT
0.456677166141693
Interpretation
Uncertain
