Gene
KWMTBOMO12583
Pre Gene Modal
BGIBMGA001436
Annotation
PREDICTED:_leucine-rich_repeat_protein_1_[Plutella_xylostella]
Full name
Leucine-rich repeat protein 1
Alternative Name
4-1BB-mediated-signaling molecule
4-1BBlrr
LRR-repeat protein 1
Peptidylprolyl isomerase-like 5
4-1BBlrr
LRR-repeat protein 1
Peptidylprolyl isomerase-like 5
Location in the cell
Extracellular Reliability : 1.735
Sequence
CDS
ATGGCCAACTTGTCTGAACTACATTTAGCAAACAATCAACTTGGTGTAAGAGGAGTAGTTGATTGGCGTTGGTTACTAGGACCTCAGATTACAAAAACTTTAAAATTATTAGATCTCACTAGTAATAAATTGGGACATCTTCCAAAAGCCATTTGGAAACTAGAAAAATTGGTAACCCTAAAAGCAAATGATAATATGATTTCAAGACTACCTGCAACTATTGGACGCATTTCCACGTTAAGATATTTCACAATCTCTAGCAATGAACTGCAATCTTTGCCATGTAGCCTAATGCAATGTAGACTGGAATACATTGACATATCCTCAAATAAATTTGACAACAAGCAAAATAATTCTACGTCAGATCAATATTCTCCGTGGCAATTTTATGTCGGAAGTTTAGTACATCTTAGTGCAAAAATAATATTAAAACATAAAATACATTATGCATCCAACATAATTCCCTGGACACTTGTTGAGTTTCTGGACAATGCTAACATGTGTGTGTGTGGAGCACCAGTAGTAAATTACACACATTCAATAAATAAAGAATATGACCTCAAAGACTATTTTAGAACTGTAGTTTTTAATAACAATCTCAGTGTGGTTGAATTTGAGTGCTATTTTTGCTCACCCAAGTGTTTCCTAAAACATTAG
Protein
MANLSELHLANNQLGVRGVVDWRWLLGPQITKTLKLLDLTSNKLGHLPKAIWKLEKLVTLKANDNMISRLPATIGRISTLRYFTISSNELQSLPCSLMQCRLEYIDISSNKFDNKQNNSTSDQYSPWQFYVGSLVHLSAKIILKHKIHYASNIIPWTLVEFLDNANMCVCGAPVVNYTHSINKEYDLKDYFRTVVFNNNLSVVEFECYFCSPKCFLKH
Summary
Description
May negatively regulate the 4-1BB-mediated signaling cascades which result in the activation of NK-kappaB and JNK1. Probable substrate recognition subunit of an ECS (Elongin BC-CUL2/5-SOCS-box protein) E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins.
Subunit
Interacts with the cytoplasmic domain of TNFRSF9. Component of the probable ECS(LRR1) E3 ubiquitin-protein ligase complex which contains CUL2, RBX1, Elongin BC complex and LRR1. Interacts with CUL2, RBX1, ELOB and ELOC.
Keywords
Alternative splicing
Complete proteome
Leucine-rich repeat
Polymorphism
Reference proteome
Repeat
Ubl conjugation pathway
Feature
chain Leucine-rich repeat protein 1
splice variant In isoform 2.
sequence variant In dbSNP:rs17121605.
splice variant In isoform 2.
sequence variant In dbSNP:rs17121605.
Uniprot
H9IW05
A0A2A4K3W5
A0A194Q4U4
A0A3S2LE53
A0A212EWM4
A0A194RQ00
+ More
A0A2H1VLW0 A0A0L7LRC7 S4PB46 A0A0C9PX45 A0A2A3E750 A0A154PPU4 A0A232EKD9 A0A0T6BFC1 D6WTS9 K7J1K9 A0A0L0C8R2 N6TQT3 U4UIW4 A0A1Y1MNK0 A0A1A9WZU5 A0A2J7RIX6 A0A1I8MC35 E2C6Q4 A0A1B0B8Y0 A0A1A9UTK6 A0A1B0FK91 A0A1A9Y5R8 A0A1A9ZPW7 A0A0N0BG93 A0A151WVY8 A0A067RED3 A0A182G3B1 G3VCB4 A0A1B6JYR3 K1PE70 A0A1B6HA38 A0A401PX57 A0A026VSH3 A0A195EBV8 A0A034W797 A0A1S3EUT6 A0A023EZ12 A0A182URA7 I3M8W2 A0A0K8W005 F6SP71 A0A1B0D5S6 A0A1B6H0U5 A0A401S4L6 A0A151IM93 E9ILT8 A0A2K6GNS9 A0A286XRL5 Q0IEE8 A0A2K5IBL3 A7S7S6 A0A0A1WQM5 D3ZE86 G5BW64 E1ZZ28 G1RMR8 A0A0G2KDW9 W8BN63 W5N046 A0A146KPM5 A0A2K5XAR6 A0A0A9YMT3 Q96L50 A0A182U610 A0A2R9B8G0 H2Q888 A0A1W4WA27 A0A1D5QNU9 W5JNV3 H0X8L9 A0A2T7NZR4 A0A096NUT2 G7MXP6 Q6NYX1 K9KAR3 A0A1A6GE12 G3R676 H9Z559 G7PA70 A0A2K6DGN5 A0A2K6PBC2 A0A2K5PL81 A0A2K6UZE3 A0A2K6KAU6 A0A2K5NQ99 A0A2K6PBA1 F7HCJ4 G2HFU2 A0A2K5DGF9 F4WNH6 L5JYG2 A0A182MTC1 A0A0D9RRX6
A0A2H1VLW0 A0A0L7LRC7 S4PB46 A0A0C9PX45 A0A2A3E750 A0A154PPU4 A0A232EKD9 A0A0T6BFC1 D6WTS9 K7J1K9 A0A0L0C8R2 N6TQT3 U4UIW4 A0A1Y1MNK0 A0A1A9WZU5 A0A2J7RIX6 A0A1I8MC35 E2C6Q4 A0A1B0B8Y0 A0A1A9UTK6 A0A1B0FK91 A0A1A9Y5R8 A0A1A9ZPW7 A0A0N0BG93 A0A151WVY8 A0A067RED3 A0A182G3B1 G3VCB4 A0A1B6JYR3 K1PE70 A0A1B6HA38 A0A401PX57 A0A026VSH3 A0A195EBV8 A0A034W797 A0A1S3EUT6 A0A023EZ12 A0A182URA7 I3M8W2 A0A0K8W005 F6SP71 A0A1B0D5S6 A0A1B6H0U5 A0A401S4L6 A0A151IM93 E9ILT8 A0A2K6GNS9 A0A286XRL5 Q0IEE8 A0A2K5IBL3 A7S7S6 A0A0A1WQM5 D3ZE86 G5BW64 E1ZZ28 G1RMR8 A0A0G2KDW9 W8BN63 W5N046 A0A146KPM5 A0A2K5XAR6 A0A0A9YMT3 Q96L50 A0A182U610 A0A2R9B8G0 H2Q888 A0A1W4WA27 A0A1D5QNU9 W5JNV3 H0X8L9 A0A2T7NZR4 A0A096NUT2 G7MXP6 Q6NYX1 K9KAR3 A0A1A6GE12 G3R676 H9Z559 G7PA70 A0A2K6DGN5 A0A2K6PBC2 A0A2K5PL81 A0A2K6UZE3 A0A2K6KAU6 A0A2K5NQ99 A0A2K6PBA1 F7HCJ4 G2HFU2 A0A2K5DGF9 F4WNH6 L5JYG2 A0A182MTC1 A0A0D9RRX6
Pubmed
19121390
26354079
22118469
26227816
23622113
28648823
+ More
18362917 19820115 20075255 26108605 23537049 28004739 25315136 20798317 24845553 26483478 21709235 22992520 30297745 24508170 30249741 25348373 25474469 17495919 21282665 21993624 17510324 17615350 25830018 15057822 15632090 21993625 23594743 24495485 26823975 25401762 11804328 14702039 15489334 15601820 22722832 16136131 17431167 20920257 23761445 22002653 22398555 25319552 25362486 21484476 21719571 23258410
18362917 19820115 20075255 26108605 23537049 28004739 25315136 20798317 24845553 26483478 21709235 22992520 30297745 24508170 30249741 25348373 25474469 17495919 21282665 21993624 17510324 17615350 25830018 15057822 15632090 21993625 23594743 24495485 26823975 25401762 11804328 14702039 15489334 15601820 22722832 16136131 17431167 20920257 23761445 22002653 22398555 25319552 25362486 21484476 21719571 23258410
EMBL
BABH01006274
NWSH01000211
PCG78360.1
KQ459585
KPI98430.1
RSAL01000188
+ More
RVE44756.1 AGBW02011934 OWR45892.1 KQ459896 KPJ19390.1 ODYU01003259 SOQ41776.1 JTDY01000265 KOB78005.1 GAIX01006117 JAA86443.1 GBYB01006078 JAG75845.1 KZ288347 PBC27577.1 KQ435012 KZC13767.1 NNAY01003809 OXU18827.1 LJIG01000903 KRT86037.1 KQ971352 EFA06739.1 JRES01000842 KNC27784.1 APGK01002320 APGK01058328 KB741288 KB734730 ENN70671.1 ENN83517.1 KB632322 ERL92338.1 GEZM01027712 JAV86638.1 NEVH01003023 PNF40776.1 GL453161 EFN76366.1 JXJN01010105 CCAG010009529 KQ435789 KOX74442.1 KQ982706 KYQ51851.1 KK852747 KDR17246.1 JXUM01141261 KQ569212 KXJ68717.1 AEFK01043639 GECU01003386 JAT04321.1 JH816535 EKC22182.1 GECU01036247 JAS71459.1 BFAA01011634 GCB77727.1 KK108417 QOIP01000014 EZA46708.1 RLU14723.1 KQ979074 KYN22710.1 GAKP01008368 JAC50584.1 GBBI01004746 JAC13966.1 AGTP01067343 GDHF01007893 JAI44421.1 AJVK01025572 AJVK01025573 GECZ01001476 JAS68293.1 BEZZ01000084 GCC25328.1 KQ977063 KYN06000.1 GL764129 EFZ18507.1 AAKN02027600 CH477695 EAT37151.1 DS469594 EDO40291.1 GBXI01013341 JAD00951.1 AABR07064716 CH473947 EDM03499.1 JH172194 GEBF01003014 EHB13524.1 JAO00619.1 GL435281 EFN73547.1 ADFV01039962 ADFV01039963 ADFV01039964 ADFV01039965 ADFV01039966 CABZ01079358 CABZ01101914 GAMC01004065 JAC02491.1 AHAT01003708 GDHC01021659 JAP96969.1 GBHO01011196 GBRD01002126 JAG32408.1 JAG63695.1 AY052405 BX248298 AK293156 CH471078 BC030142 BC093697 BC112241 BC139921 AJFE02112253 AJFE02112254 AJFE02112255 AJFE02112256 AJFE02112257 AJFE02112258 AACZ04059603 GABC01006043 GABF01004597 GABD01007302 GABE01008924 NBAG03000214 JAA05295.1 JAA17548.1 JAA25798.1 JAA35815.1 PNI83274.1 JSUE03039692 JSUE03039693 ADMH02001019 ETN64429.1 AAQR03088013 AAQR03088014 PZQS01000008 PVD26660.1 AHZZ02032136 AHZZ02032137 AHZZ02032138 CM001259 EHH27854.1 BC066431 AAH66431.1 JL621978 AEP99538.1 LZPO01097182 OBS64486.1 CABD030092220 CABD030092221 JU474271 AFH31075.1 CM001282 EHH63595.1 AK305606 BAK62600.1 GL888237 EGI64169.1 KB031080 ELK03546.1 AXCM01001019 AQIB01058984
RVE44756.1 AGBW02011934 OWR45892.1 KQ459896 KPJ19390.1 ODYU01003259 SOQ41776.1 JTDY01000265 KOB78005.1 GAIX01006117 JAA86443.1 GBYB01006078 JAG75845.1 KZ288347 PBC27577.1 KQ435012 KZC13767.1 NNAY01003809 OXU18827.1 LJIG01000903 KRT86037.1 KQ971352 EFA06739.1 JRES01000842 KNC27784.1 APGK01002320 APGK01058328 KB741288 KB734730 ENN70671.1 ENN83517.1 KB632322 ERL92338.1 GEZM01027712 JAV86638.1 NEVH01003023 PNF40776.1 GL453161 EFN76366.1 JXJN01010105 CCAG010009529 KQ435789 KOX74442.1 KQ982706 KYQ51851.1 KK852747 KDR17246.1 JXUM01141261 KQ569212 KXJ68717.1 AEFK01043639 GECU01003386 JAT04321.1 JH816535 EKC22182.1 GECU01036247 JAS71459.1 BFAA01011634 GCB77727.1 KK108417 QOIP01000014 EZA46708.1 RLU14723.1 KQ979074 KYN22710.1 GAKP01008368 JAC50584.1 GBBI01004746 JAC13966.1 AGTP01067343 GDHF01007893 JAI44421.1 AJVK01025572 AJVK01025573 GECZ01001476 JAS68293.1 BEZZ01000084 GCC25328.1 KQ977063 KYN06000.1 GL764129 EFZ18507.1 AAKN02027600 CH477695 EAT37151.1 DS469594 EDO40291.1 GBXI01013341 JAD00951.1 AABR07064716 CH473947 EDM03499.1 JH172194 GEBF01003014 EHB13524.1 JAO00619.1 GL435281 EFN73547.1 ADFV01039962 ADFV01039963 ADFV01039964 ADFV01039965 ADFV01039966 CABZ01079358 CABZ01101914 GAMC01004065 JAC02491.1 AHAT01003708 GDHC01021659 JAP96969.1 GBHO01011196 GBRD01002126 JAG32408.1 JAG63695.1 AY052405 BX248298 AK293156 CH471078 BC030142 BC093697 BC112241 BC139921 AJFE02112253 AJFE02112254 AJFE02112255 AJFE02112256 AJFE02112257 AJFE02112258 AACZ04059603 GABC01006043 GABF01004597 GABD01007302 GABE01008924 NBAG03000214 JAA05295.1 JAA17548.1 JAA25798.1 JAA35815.1 PNI83274.1 JSUE03039692 JSUE03039693 ADMH02001019 ETN64429.1 AAQR03088013 AAQR03088014 PZQS01000008 PVD26660.1 AHZZ02032136 AHZZ02032137 AHZZ02032138 CM001259 EHH27854.1 BC066431 AAH66431.1 JL621978 AEP99538.1 LZPO01097182 OBS64486.1 CABD030092220 CABD030092221 JU474271 AFH31075.1 CM001282 EHH63595.1 AK305606 BAK62600.1 GL888237 EGI64169.1 KB031080 ELK03546.1 AXCM01001019 AQIB01058984
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000007151
UP000053240
+ More
UP000037510 UP000242457 UP000076502 UP000215335 UP000007266 UP000002358 UP000037069 UP000019118 UP000030742 UP000091820 UP000235965 UP000095301 UP000008237 UP000092460 UP000078200 UP000092444 UP000092443 UP000092445 UP000053105 UP000075809 UP000027135 UP000069940 UP000249989 UP000007648 UP000005408 UP000288216 UP000053097 UP000279307 UP000078492 UP000081671 UP000075903 UP000005215 UP000002280 UP000092462 UP000287033 UP000078542 UP000233160 UP000005447 UP000008820 UP000233080 UP000001593 UP000002494 UP000006813 UP000000311 UP000001073 UP000000437 UP000018468 UP000233140 UP000005640 UP000075902 UP000240080 UP000002277 UP000192223 UP000006718 UP000000673 UP000005225 UP000245119 UP000028761 UP000092124 UP000001519 UP000009130 UP000233120 UP000233200 UP000233040 UP000233220 UP000233180 UP000233060 UP000008225 UP000233020 UP000007755 UP000010552 UP000075883 UP000029965
UP000037510 UP000242457 UP000076502 UP000215335 UP000007266 UP000002358 UP000037069 UP000019118 UP000030742 UP000091820 UP000235965 UP000095301 UP000008237 UP000092460 UP000078200 UP000092444 UP000092443 UP000092445 UP000053105 UP000075809 UP000027135 UP000069940 UP000249989 UP000007648 UP000005408 UP000288216 UP000053097 UP000279307 UP000078492 UP000081671 UP000075903 UP000005215 UP000002280 UP000092462 UP000287033 UP000078542 UP000233160 UP000005447 UP000008820 UP000233080 UP000001593 UP000002494 UP000006813 UP000000311 UP000001073 UP000000437 UP000018468 UP000233140 UP000005640 UP000075902 UP000240080 UP000002277 UP000192223 UP000006718 UP000000673 UP000005225 UP000245119 UP000028761 UP000092124 UP000001519 UP000009130 UP000233120 UP000233200 UP000233040 UP000233220 UP000233180 UP000233060 UP000008225 UP000233020 UP000007755 UP000010552 UP000075883 UP000029965
Interpro
Gene 3D
ProteinModelPortal
H9IW05
A0A2A4K3W5
A0A194Q4U4
A0A3S2LE53
A0A212EWM4
A0A194RQ00
+ More
A0A2H1VLW0 A0A0L7LRC7 S4PB46 A0A0C9PX45 A0A2A3E750 A0A154PPU4 A0A232EKD9 A0A0T6BFC1 D6WTS9 K7J1K9 A0A0L0C8R2 N6TQT3 U4UIW4 A0A1Y1MNK0 A0A1A9WZU5 A0A2J7RIX6 A0A1I8MC35 E2C6Q4 A0A1B0B8Y0 A0A1A9UTK6 A0A1B0FK91 A0A1A9Y5R8 A0A1A9ZPW7 A0A0N0BG93 A0A151WVY8 A0A067RED3 A0A182G3B1 G3VCB4 A0A1B6JYR3 K1PE70 A0A1B6HA38 A0A401PX57 A0A026VSH3 A0A195EBV8 A0A034W797 A0A1S3EUT6 A0A023EZ12 A0A182URA7 I3M8W2 A0A0K8W005 F6SP71 A0A1B0D5S6 A0A1B6H0U5 A0A401S4L6 A0A151IM93 E9ILT8 A0A2K6GNS9 A0A286XRL5 Q0IEE8 A0A2K5IBL3 A7S7S6 A0A0A1WQM5 D3ZE86 G5BW64 E1ZZ28 G1RMR8 A0A0G2KDW9 W8BN63 W5N046 A0A146KPM5 A0A2K5XAR6 A0A0A9YMT3 Q96L50 A0A182U610 A0A2R9B8G0 H2Q888 A0A1W4WA27 A0A1D5QNU9 W5JNV3 H0X8L9 A0A2T7NZR4 A0A096NUT2 G7MXP6 Q6NYX1 K9KAR3 A0A1A6GE12 G3R676 H9Z559 G7PA70 A0A2K6DGN5 A0A2K6PBC2 A0A2K5PL81 A0A2K6UZE3 A0A2K6KAU6 A0A2K5NQ99 A0A2K6PBA1 F7HCJ4 G2HFU2 A0A2K5DGF9 F4WNH6 L5JYG2 A0A182MTC1 A0A0D9RRX6
A0A2H1VLW0 A0A0L7LRC7 S4PB46 A0A0C9PX45 A0A2A3E750 A0A154PPU4 A0A232EKD9 A0A0T6BFC1 D6WTS9 K7J1K9 A0A0L0C8R2 N6TQT3 U4UIW4 A0A1Y1MNK0 A0A1A9WZU5 A0A2J7RIX6 A0A1I8MC35 E2C6Q4 A0A1B0B8Y0 A0A1A9UTK6 A0A1B0FK91 A0A1A9Y5R8 A0A1A9ZPW7 A0A0N0BG93 A0A151WVY8 A0A067RED3 A0A182G3B1 G3VCB4 A0A1B6JYR3 K1PE70 A0A1B6HA38 A0A401PX57 A0A026VSH3 A0A195EBV8 A0A034W797 A0A1S3EUT6 A0A023EZ12 A0A182URA7 I3M8W2 A0A0K8W005 F6SP71 A0A1B0D5S6 A0A1B6H0U5 A0A401S4L6 A0A151IM93 E9ILT8 A0A2K6GNS9 A0A286XRL5 Q0IEE8 A0A2K5IBL3 A7S7S6 A0A0A1WQM5 D3ZE86 G5BW64 E1ZZ28 G1RMR8 A0A0G2KDW9 W8BN63 W5N046 A0A146KPM5 A0A2K5XAR6 A0A0A9YMT3 Q96L50 A0A182U610 A0A2R9B8G0 H2Q888 A0A1W4WA27 A0A1D5QNU9 W5JNV3 H0X8L9 A0A2T7NZR4 A0A096NUT2 G7MXP6 Q6NYX1 K9KAR3 A0A1A6GE12 G3R676 H9Z559 G7PA70 A0A2K6DGN5 A0A2K6PBC2 A0A2K5PL81 A0A2K6UZE3 A0A2K6KAU6 A0A2K5NQ99 A0A2K6PBA1 F7HCJ4 G2HFU2 A0A2K5DGF9 F4WNH6 L5JYG2 A0A182MTC1 A0A0D9RRX6
PDB
4U06
E-value=1.76569e-07,
Score=129
Ontologies
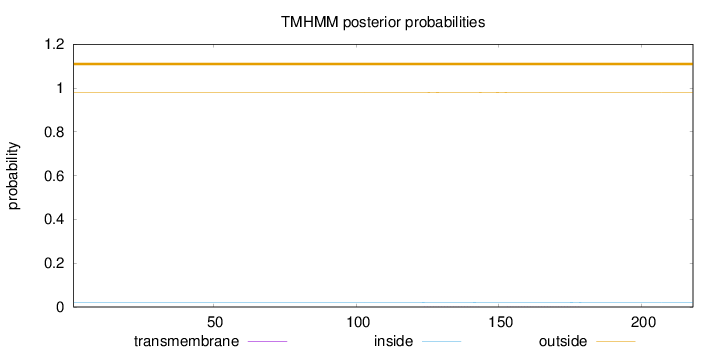
Topology
Length:
218
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0341
Exp number, first 60 AAs:
0.00062
Total prob of N-in:
0.02026
outside
1 - 218
Population Genetic Test Statistics
Pi
213.113012
Theta
198.741727
Tajima's D
0.199723
CLR
0
CSRT
0.422728863556822
Interpretation
Uncertain
