Pre Gene Modal
BGIBMGA001538
Annotation
elongator_complex_protein_1_[Bombyx_mori]
Full name
Elongator complex protein 1
Alternative Name
IkappaB kinase complex-associated protein
Location in the cell
Cytoplasmic Reliability : 2.098 Nuclear Reliability : 1.756
Sequence
CDS
ATGAAGAATCTGCAACTTTGGGATATTTCTATCAGAGATTTTTGTGTTCATTTTGAAGAGAAAGCTGTAATATGTCATGGCCGTGGCGACGGAGGCTCATCAAGTGATATATTTGTTTGTGGAAATAACCTTATTTTATGCAATTTTGATGAAAATTGCCAGAAAAAGTGGTTTAAGGATGTTTCTGAATACATATCTCCTGAAAATAGCCCTGTGAATATATGTTACCTGTCGCTTCAAAACTCACTATGCATCGGTTTTGCGAATGGAGAGTTATTAGCCATAGGTGATTCCGGTGTCACGTGTGAAACAGTTGGAATCTTCAATACTGGATTACTGGCCATGGAATGGAGTCCTGATCAAGAATTATTAGCAATTGTCACTAAAGAAATGCACATGGTCCTGTTGTCTTGCACTTATGATCCAATTAATGAAATTAAATTGCACAATCAAGAGTTTGGTGAGAAGCAGTTTATAACTGTTGGGTGGGGCAAAAAAGAAACACAGTTTCATGGATCTGAAGGGAAACAGGCAGCTAAAATAAAAAATGACATCGTTTCAGACGACTCGACTGCCAGTGATAAAGTTAAAATTACCTGGAGAGGAGATGGTAATTTGTATGCCATTGGTTTTGCAATGGATGGGCTTAGACATTTTAAAGTTTTTGACAGAGAAGGTCATTTACTATACACAAGTGAAAAGCAAAAAGGTCTAGAAGACAATATTGTATGGCGTCCAAGTGGGAATGTAATCACGACAACACAAAAAATTGAAAACCAGTATAAAGTAACATTTTTTGAAAAGAATGGTCTCAAACATAGAGAGTTCAGTATTCCAGTAGAACATGGTACATATGTTGAAAATATCATGTGGAGCTCGGACTCTGAAATATTAACTCTGCAATGTAAAGATACAGAAACAAACACACAAAAAGTATTATTATACACAACATCAAATTACCATTGGTATTTGAAACAAACATTGTTGTTTAGAAGTAATCAACGCATCAATAAAATCATCTGGGATAATGATTTTGACATATCAAACAACAAAAAAATGCATATTATTTTACAAAATGGATCTTATTTGATATATTCATGGATTTGGAATATTGATCACAGTAAAAGTAATATTGATAATGATGATGCTGTTGTAGTTGCTATTGATGGAGATAAATTATTGATCACTGGATTTAGACAAACTGTAGTACCTCCACCAATGGCATCACTTGAAATAAAATTGGAATCTACTGCACAGGCAATATGTTTTGCACCAAAAAACGAAAATGTTAACCCAAATTCATTCTTTGTAATTACAGTAGATAACAAATTACTTTTTTACTCACAAAAAGAGAAACATCCACTCAATTATGAAGCATATAACTCTCAGCAATTTGACAAATCTGATTTTCCATTCCAATACCATCACTGGCTCTGGGTAGACTCTCAAACAATAGTGTGTGCTATGACAGATGAAAAGTCAACCTCTGTTATAGAACTGACTATGGTGAATGACAAACTTGTTAAAAGTAACAGTGTACATGTAAGAGGTGTTGTGACACAAATACAAGCACATCCTACAAATAAATCTTTACTATTCTTGCAGTTTATAACTGGTCATATACATAAATACAATATTGGTGGTTTTCTTGAAAATACTAATATATTATTTAAAGTACCATGTCCCAGGTTTGGTATATTGCCTATTGAAGACTCTTTGCATTTCATTGGACTTTCACACAAGGGCCATTTGTTTATTGATAATGTACAGGTTCTCAGTAATGTTAGTTCTTTCTTCATACATACGGATTTCTTGTTGTTGACTACCTTACAACATTTGTTATTGTGTGTGGAAAAGAACAAACTGGGTTTGAAAGCTCTGATGGAATACCAAACAAATGAATCTGATTATGTATATAAAAGAAAAATTGAACGAGGTGCCAAATTAGTAATTATAGTACCAAATGATACTAGAACTATTTTGCAGATGCCAAGAGGCAATATAGAAGCAATACAACCCAGACCTCTGTCTCTTAAAATTATTGGAAAATATCTGGATAACTTGAAGTACTATGAAGCATTTGATTTGATGAGGAAACAGAGAATCAATTTAAACCTTATTTTTGACCACAATCCAAAAAAGTTTATAGCGAATATTGACACATTTTTACATTCCATTAAAAACAATTCTTGGCTGAATCTATTTTTATCTGATTTAGAAAATATAGATGTTACAAAAACAATGTATTCAAGTATAAATTATTATGCTGAAAGACCTGCTGTAACTGATGAAATTTCAAGAAAGATTGATATTGTTTGTGAAATGTTTCGAGCTCATATAGAAAAAAGATCAGATAAGGCAACTAAAATCTTACCACTTCTCACAACTTATGTAAAGAAAAATACTGTAGATGACTTGCAAAAAGCCTTGGAAATAATTAAAGGTTTAAAAAAACAAGAGACATCTGGTGAGAAAATTCCTGTAAGTTCTGATGAAGCCCTCAAGTATTTGTTGTATATGGTAGATGTTACCCAATTATTTGACATTGCACTCGGGATGTATGATTTTGAATTAGTTTTGCTAGTAGCAACTAAGTCACAGAAAGATCCAAAAGAATACATTCCTATGCTAAATGAGTTAAATGAAATGGACGAAAATTACAAACGATTTACAATAAATAAGCAATTGAAAAGATTTGATAAAGCAGTGCAATCTCTTGTACTGTGCGGTCCAACACGTCACTGTGAACTGAAAACTTTCGTAAAATATCATAGTTTGTATCAAGAGGCGTTGAAACATTTTTCTTTTGAAGAAGAAATTTTCAGGGAAATTTCTGAAGATTATGGACAACACTTGAAATTGAAGAAATACTACACAGAAGCTGCTATTATTTATGAAAGAGCTAATAATAATGACAAAGCTATTGAATGTTACAAAGAAGCTCTAGAATGGGAATTGGCGATCAAACTAGCCTATTTTTGGCCAAAGGCACAATTCAAAGTGTTGTGCTGGGAGCTAGTAACTGCACTGAAAGAAGAAAAACGTCACGAAGAAGCTCTAATCATTTTAGAACAATTCTACGGCGATCCTGAGGAGTGCATTAGTTATGCGGTTGAAACTAGTCACTACAAAAAAGCTTTACGTCTTTGTTCGCTATATGATAAACTACAATTGAAAGAGGAACGCATACTACCTGCTCTCCTTGAAGAATACCAAAATATGACAGATTTAATAGAAACTAATCGGAGCACATTTTTAAAACATCGTGAACGACTTTTTCACGTGAGAAATATAAAGAGGGACAATCCAGTTGATCTCTACGATATATACACAAATAAAGATGCGGATCTATATTCAGATGCCGGGAGTACTTTGGCGTCGTCTTCTAGAGGTTCGAGCCGTTCATACCGTTCAAGCAAGAACCGAAGGAAACACGAACGTAAAATAGCATCTCTCAAAGAAGGGTCGCAGTATGAAGACGTTGGATTAATTATTGCACTACACTGCCTCATTACATCGACGTTTGACTTGAGAAATCATGTTAAAGATCTTACTGTTGGGTTGATTTGCTTTAATATGGACAAAGAAGCATTCATATTGCAGAAGGCATTAGAGAAGTTATTAAATGAAATGAAAGACAGTTTTAAAGATATTTGGACAAATGATTTTATGTTAGAAGCTACAAATGCAACTATCACAGCACATAATATATCAGAGGGATCTAGTGTGTTACCACCAGGCATTGCTTCATTAGAACTACATTTCAGAATACCACCTGTAATACAGGAGATTAATTGGAAATTAGAAGGTCTCAGTTAA
Protein
MKNLQLWDISIRDFCVHFEEKAVICHGRGDGGSSSDIFVCGNNLILCNFDENCQKKWFKDVSEYISPENSPVNICYLSLQNSLCIGFANGELLAIGDSGVTCETVGIFNTGLLAMEWSPDQELLAIVTKEMHMVLLSCTYDPINEIKLHNQEFGEKQFITVGWGKKETQFHGSEGKQAAKIKNDIVSDDSTASDKVKITWRGDGNLYAIGFAMDGLRHFKVFDREGHLLYTSEKQKGLEDNIVWRPSGNVITTTQKIENQYKVTFFEKNGLKHREFSIPVEHGTYVENIMWSSDSEILTLQCKDTETNTQKVLLYTTSNYHWYLKQTLLFRSNQRINKIIWDNDFDISNNKKMHIILQNGSYLIYSWIWNIDHSKSNIDNDDAVVVAIDGDKLLITGFRQTVVPPPMASLEIKLESTAQAICFAPKNENVNPNSFFVITVDNKLLFYSQKEKHPLNYEAYNSQQFDKSDFPFQYHHWLWVDSQTIVCAMTDEKSTSVIELTMVNDKLVKSNSVHVRGVVTQIQAHPTNKSLLFLQFITGHIHKYNIGGFLENTNILFKVPCPRFGILPIEDSLHFIGLSHKGHLFIDNVQVLSNVSSFFIHTDFLLLTTLQHLLLCVEKNKLGLKALMEYQTNESDYVYKRKIERGAKLVIIVPNDTRTILQMPRGNIEAIQPRPLSLKIIGKYLDNLKYYEAFDLMRKQRINLNLIFDHNPKKFIANIDTFLHSIKNNSWLNLFLSDLENIDVTKTMYSSINYYAERPAVTDEISRKIDIVCEMFRAHIEKRSDKATKILPLLTTYVKKNTVDDLQKALEIIKGLKKQETSGEKIPVSSDEALKYLLYMVDVTQLFDIALGMYDFELVLLVATKSQKDPKEYIPMLNELNEMDENYKRFTINKQLKRFDKAVQSLVLCGPTRHCELKTFVKYHSLYQEALKHFSFEEEIFREISEDYGQHLKLKKYYTEAAIIYERANNNDKAIECYKEALEWELAIKLAYFWPKAQFKVLCWELVTALKEEKRHEEALIILEQFYGDPEECISYAVETSHYKKALRLCSLYDKLQLKEERILPALLEEYQNMTDLIETNRSTFLKHRERLFHVRNIKRDNPVDLYDIYTNKDADLYSDAGSTLASSSRGSSRSYRSSKNRRKHERKIASLKEGSQYEDVGLIIALHCLITSTFDLRNHVKDLTVGLICFNMDKEAFILQKALEKLLNEMKDSFKDIWTNDFMLEATNATITAHNISEGSSVLPPGIASLELHFRIPPVIQEINWKLEGLS
Summary
Description
Acts as subunit of the RNA polymerase II elongator complex, which is a histone acetyltransferase component of the RNA polymerase II (Pol II) holoenzyme and is involved in transcriptional elongation.
May act as a scaffold protein that may assemble active IKK-MAP3K14 complexes (IKKA, IKKB and MAP3K14/NIK).
Acts as subunit of the RNA polymerase II elongator complex, which is a histone acetyltransferase component of the RNA polymerase II (Pol II) holoenzyme and is involved in transcriptional elongation. Elongator may play a role in chromatin remodeling and is involved in acetylation of histones H3 and probably H4. Involved in cell migration (By similarity). Involved in neurogenesis (By similarity). Regulates the migration and branching of projection neurons in the developing cerebral cortex, through a process depending on alpha-tubulin acetylation (By similarity).
May act as a scaffold protein that may assemble active IKK-MAP3K14 complexes (IKKA, IKKB and MAP3K14/NIK).
Acts as subunit of the RNA polymerase II elongator complex, which is a histone acetyltransferase component of the RNA polymerase II (Pol II) holoenzyme and is involved in transcriptional elongation. Elongator may play a role in chromatin remodeling and is involved in acetylation of histones H3 and probably H4. Involved in cell migration (By similarity). Involved in neurogenesis (By similarity). Regulates the migration and branching of projection neurons in the developing cerebral cortex, through a process depending on alpha-tubulin acetylation (By similarity).
Subunit
Interacts preferentially with MAP3K14/NIK followed by IKK-alpha and IKK-beta. Component of the RNA polymerase II elongator complex (Elongator), which consists of ELP1, STIP1/ELP2, ELP3, ELP4, ELP5 and ELP6. Elongator associates with the C-terminal domain (CTD) of Pol II largest subunit. Interacts with ELP3 (By similarity).
Similarity
Belongs to the ELP1/IKA1 family.
Keywords
Complete proteome
Cytoplasm
Nucleus
Phosphoprotein
Reference proteome
Transcription
Transcription regulation
Feature
chain Elongator complex protein 1
Uniprot
D9N4J5
A0A2A4K266
A0A1E1VXL1
A0A2H1VLT3
A0A437B3L3
A0A194RTE8
+ More
A0A194Q0A8 A0A0L7LRH3 A0A2J7R223 A0A067R977 A0A232FE73 K7ISC0 A0A401NYF1 A0A1V4J6J6 A0A1S3H0H9 A0A2I4D024 A0A3B3QPX9 A0A2I0M5E6 A0A218UIA2 A0A0M8ZST8 A0A0C9R4C0 A0A1L8HN20 A0A3Q3VQI1 A0A3B4WUV3 A0A091QZ03 A0A151NEJ8 K7F2Q5 A0A151NE03 A0A3Q3LKD3 A0A0R4IJ07 A0A0L7RCB5 K7F2Q8 A0A3Q0G9N0 A0A1A7W7B7 A0A3B3CMQ8 I3M7Y6 A0A2U9C4V4 A0A3P9BHT6 V4A9R2 A0A3Q2X553 A0A3P8PZ67 A0A1W5AQ41 A0A3B3YXS1 A0A3Q2X3Y5 A0A087Y518 A0A3Q4HG62 K1Q1G2 A0A3B4F1K3 A0A3Q3IJB3 I3IZA3 A0A3P8W0K3 A0A3Q3F7Y9 A0A3P8VXP7 A0A2A3EJL4 A0A087ZQT9 A0A3P9NNM2 A0A452G121 A0A3P8RRC5 A0A310SHL8 A0A1A8J9B7 A0A3P9L654 A0A154PLA7 M4AXK8 A0A146X6E4 A0A1A8AKL9 A0A1A8VC89 W5PB34 A0A3P9NNT6 G3PX47 U3JES9 H2M688 A0A096MAM9 A0A250YM53 A0A060W5N3 A0A1A8R1T5 A0A1A8R7B7 A0A1A8M2M4 A0A1A8LYF9 A0A1S3FQ83 A0A2B4T0H9 H3D601 A0A146NQN2 A0A1A8FKJ7 A0A452UL10 A0A384DH79 A0A2I2U2Q5 A0A3Q7VLE7 Q8WND5 B2RY98 A0A2K5ECS6 A0A182G4H9 A0A1A8GIZ6 A0A1S4EXL4 A0A2K5J1X1 A0A0K8RU22 Q17MG4 F1Q372 H0V6K7 L5LEY5
A0A194Q0A8 A0A0L7LRH3 A0A2J7R223 A0A067R977 A0A232FE73 K7ISC0 A0A401NYF1 A0A1V4J6J6 A0A1S3H0H9 A0A2I4D024 A0A3B3QPX9 A0A2I0M5E6 A0A218UIA2 A0A0M8ZST8 A0A0C9R4C0 A0A1L8HN20 A0A3Q3VQI1 A0A3B4WUV3 A0A091QZ03 A0A151NEJ8 K7F2Q5 A0A151NE03 A0A3Q3LKD3 A0A0R4IJ07 A0A0L7RCB5 K7F2Q8 A0A3Q0G9N0 A0A1A7W7B7 A0A3B3CMQ8 I3M7Y6 A0A2U9C4V4 A0A3P9BHT6 V4A9R2 A0A3Q2X553 A0A3P8PZ67 A0A1W5AQ41 A0A3B3YXS1 A0A3Q2X3Y5 A0A087Y518 A0A3Q4HG62 K1Q1G2 A0A3B4F1K3 A0A3Q3IJB3 I3IZA3 A0A3P8W0K3 A0A3Q3F7Y9 A0A3P8VXP7 A0A2A3EJL4 A0A087ZQT9 A0A3P9NNM2 A0A452G121 A0A3P8RRC5 A0A310SHL8 A0A1A8J9B7 A0A3P9L654 A0A154PLA7 M4AXK8 A0A146X6E4 A0A1A8AKL9 A0A1A8VC89 W5PB34 A0A3P9NNT6 G3PX47 U3JES9 H2M688 A0A096MAM9 A0A250YM53 A0A060W5N3 A0A1A8R1T5 A0A1A8R7B7 A0A1A8M2M4 A0A1A8LYF9 A0A1S3FQ83 A0A2B4T0H9 H3D601 A0A146NQN2 A0A1A8FKJ7 A0A452UL10 A0A384DH79 A0A2I2U2Q5 A0A3Q7VLE7 Q8WND5 B2RY98 A0A2K5ECS6 A0A182G4H9 A0A1A8GIZ6 A0A1S4EXL4 A0A2K5J1X1 A0A0K8RU22 Q17MG4 F1Q372 H0V6K7 L5LEY5
Pubmed
EMBL
AB573815
BAJ14102.1
NWSH01000211
PCG78361.1
GDQN01011571
JAT79483.1
+ More
ODYU01003259 SOQ41777.1 RSAL01000188 RVE44755.1 KQ459896 KPJ19391.1 KQ459585 KPI98429.1 JTDY01000265 KOB78004.1 NEVH01008200 PNF34888.1 KK852805 KDR16130.1 NNAY01000349 OXU28995.1 BFAA01006936 GCB65885.1 LSYS01009041 OPJ67407.1 AKCR02000037 PKK24905.1 MUZQ01000284 OWK53509.1 KQ435911 KOX68903.1 GBYB01007657 JAG77424.1 CM004467 OCT97451.1 KK710106 KFQ32785.1 AKHW03003207 KYO35039.1 AGCU01154302 AGCU01154303 AGCU01154304 AGCU01154305 AGCU01154306 AGCU01154307 AGCU01154308 AGCU01154309 AGCU01154310 KYO35038.1 FP103010 KQ414616 KOC68440.1 HADW01000368 SBP01768.1 AGTP01059131 AGTP01059132 CP026255 AWP11615.1 KB200701 ESP00729.1 AYCK01022059 JH816701 EKC25194.1 AERX01009992 AERX01009993 KZ288226 PBC31890.1 LWLT01000007 KQ769179 OAD52932.1 HAED01018853 SBR05298.1 KQ434960 KZC12649.1 GCES01048635 JAR37688.1 HADY01016275 SBP54760.1 HADY01017318 HAEJ01017024 SBS57481.1 AMGL01047284 AGTO01001226 GFFW01000065 JAV44723.1 FR904405 CDQ62331.1 HAEH01014310 SBR99692.1 HAEH01014905 SBS01318.1 HAEF01010514 SBR50524.1 HAEF01010053 SBR49662.1 LSMT01000001 PFX35046.1 GCES01152284 JAQ34038.1 HAEB01013443 SBQ59970.1 AANG04004382 AF388202 BC166239 AAI66239.1 JXUM01143771 KQ569660 KXJ68558.1 HAEC01003693 SBQ71770.1 GBKC01001766 JAG44304.1 CH477206 EAT47874.1 AAEX03008035 AAKN02049922 AAKN02049923 KB112777 ELK24570.1
ODYU01003259 SOQ41777.1 RSAL01000188 RVE44755.1 KQ459896 KPJ19391.1 KQ459585 KPI98429.1 JTDY01000265 KOB78004.1 NEVH01008200 PNF34888.1 KK852805 KDR16130.1 NNAY01000349 OXU28995.1 BFAA01006936 GCB65885.1 LSYS01009041 OPJ67407.1 AKCR02000037 PKK24905.1 MUZQ01000284 OWK53509.1 KQ435911 KOX68903.1 GBYB01007657 JAG77424.1 CM004467 OCT97451.1 KK710106 KFQ32785.1 AKHW03003207 KYO35039.1 AGCU01154302 AGCU01154303 AGCU01154304 AGCU01154305 AGCU01154306 AGCU01154307 AGCU01154308 AGCU01154309 AGCU01154310 KYO35038.1 FP103010 KQ414616 KOC68440.1 HADW01000368 SBP01768.1 AGTP01059131 AGTP01059132 CP026255 AWP11615.1 KB200701 ESP00729.1 AYCK01022059 JH816701 EKC25194.1 AERX01009992 AERX01009993 KZ288226 PBC31890.1 LWLT01000007 KQ769179 OAD52932.1 HAED01018853 SBR05298.1 KQ434960 KZC12649.1 GCES01048635 JAR37688.1 HADY01016275 SBP54760.1 HADY01017318 HAEJ01017024 SBS57481.1 AMGL01047284 AGTO01001226 GFFW01000065 JAV44723.1 FR904405 CDQ62331.1 HAEH01014310 SBR99692.1 HAEH01014905 SBS01318.1 HAEF01010514 SBR50524.1 HAEF01010053 SBR49662.1 LSMT01000001 PFX35046.1 GCES01152284 JAQ34038.1 HAEB01013443 SBQ59970.1 AANG04004382 AF388202 BC166239 AAI66239.1 JXUM01143771 KQ569660 KXJ68558.1 HAEC01003693 SBQ71770.1 GBKC01001766 JAG44304.1 CH477206 EAT47874.1 AAEX03008035 AAKN02049922 AAKN02049923 KB112777 ELK24570.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000037510
UP000235965
+ More
UP000027135 UP000215335 UP000002358 UP000288216 UP000190648 UP000085678 UP000192220 UP000261540 UP000053872 UP000197619 UP000053105 UP000186698 UP000261620 UP000261360 UP000050525 UP000007267 UP000261640 UP000000437 UP000053825 UP000189705 UP000261560 UP000005215 UP000246464 UP000265160 UP000030746 UP000264840 UP000265100 UP000192224 UP000261480 UP000028760 UP000261580 UP000005408 UP000261460 UP000261600 UP000005207 UP000265120 UP000261660 UP000242457 UP000005203 UP000242638 UP000291000 UP000265080 UP000265180 UP000076502 UP000002852 UP000002356 UP000007635 UP000016665 UP000001038 UP000193380 UP000081671 UP000225706 UP000007303 UP000291021 UP000261680 UP000011712 UP000286642 UP000001811 UP000233020 UP000069940 UP000249989 UP000233080 UP000008820 UP000002254 UP000005447
UP000027135 UP000215335 UP000002358 UP000288216 UP000190648 UP000085678 UP000192220 UP000261540 UP000053872 UP000197619 UP000053105 UP000186698 UP000261620 UP000261360 UP000050525 UP000007267 UP000261640 UP000000437 UP000053825 UP000189705 UP000261560 UP000005215 UP000246464 UP000265160 UP000030746 UP000264840 UP000265100 UP000192224 UP000261480 UP000028760 UP000261580 UP000005408 UP000261460 UP000261600 UP000005207 UP000265120 UP000261660 UP000242457 UP000005203 UP000242638 UP000291000 UP000265080 UP000265180 UP000076502 UP000002852 UP000002356 UP000007635 UP000016665 UP000001038 UP000193380 UP000081671 UP000225706 UP000007303 UP000291021 UP000261680 UP000011712 UP000286642 UP000001811 UP000233020 UP000069940 UP000249989 UP000233080 UP000008820 UP000002254 UP000005447
Interpro
IPR011990
TPR-like_helical_dom_sf
+ More
IPR006849 Elp1
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR010793 Ribosomal_L37/S30
IPR019734 TPR_repeat
IPR013026 TPR-contain_dom
IPR036322 WD40_repeat_dom_sf
IPR000261 EH_dom
IPR036705 Ribosyl_crysJ1_sf
IPR017452 GPCR_Rhodpsn_7TM
IPR000276 GPCR_Rhodpsn
IPR011047 Quinoprotein_ADH-like_supfam
IPR006849 Elp1
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR010793 Ribosomal_L37/S30
IPR019734 TPR_repeat
IPR013026 TPR-contain_dom
IPR036322 WD40_repeat_dom_sf
IPR000261 EH_dom
IPR036705 Ribosyl_crysJ1_sf
IPR017452 GPCR_Rhodpsn_7TM
IPR000276 GPCR_Rhodpsn
IPR011047 Quinoprotein_ADH-like_supfam
Gene 3D
ProteinModelPortal
D9N4J5
A0A2A4K266
A0A1E1VXL1
A0A2H1VLT3
A0A437B3L3
A0A194RTE8
+ More
A0A194Q0A8 A0A0L7LRH3 A0A2J7R223 A0A067R977 A0A232FE73 K7ISC0 A0A401NYF1 A0A1V4J6J6 A0A1S3H0H9 A0A2I4D024 A0A3B3QPX9 A0A2I0M5E6 A0A218UIA2 A0A0M8ZST8 A0A0C9R4C0 A0A1L8HN20 A0A3Q3VQI1 A0A3B4WUV3 A0A091QZ03 A0A151NEJ8 K7F2Q5 A0A151NE03 A0A3Q3LKD3 A0A0R4IJ07 A0A0L7RCB5 K7F2Q8 A0A3Q0G9N0 A0A1A7W7B7 A0A3B3CMQ8 I3M7Y6 A0A2U9C4V4 A0A3P9BHT6 V4A9R2 A0A3Q2X553 A0A3P8PZ67 A0A1W5AQ41 A0A3B3YXS1 A0A3Q2X3Y5 A0A087Y518 A0A3Q4HG62 K1Q1G2 A0A3B4F1K3 A0A3Q3IJB3 I3IZA3 A0A3P8W0K3 A0A3Q3F7Y9 A0A3P8VXP7 A0A2A3EJL4 A0A087ZQT9 A0A3P9NNM2 A0A452G121 A0A3P8RRC5 A0A310SHL8 A0A1A8J9B7 A0A3P9L654 A0A154PLA7 M4AXK8 A0A146X6E4 A0A1A8AKL9 A0A1A8VC89 W5PB34 A0A3P9NNT6 G3PX47 U3JES9 H2M688 A0A096MAM9 A0A250YM53 A0A060W5N3 A0A1A8R1T5 A0A1A8R7B7 A0A1A8M2M4 A0A1A8LYF9 A0A1S3FQ83 A0A2B4T0H9 H3D601 A0A146NQN2 A0A1A8FKJ7 A0A452UL10 A0A384DH79 A0A2I2U2Q5 A0A3Q7VLE7 Q8WND5 B2RY98 A0A2K5ECS6 A0A182G4H9 A0A1A8GIZ6 A0A1S4EXL4 A0A2K5J1X1 A0A0K8RU22 Q17MG4 F1Q372 H0V6K7 L5LEY5
A0A194Q0A8 A0A0L7LRH3 A0A2J7R223 A0A067R977 A0A232FE73 K7ISC0 A0A401NYF1 A0A1V4J6J6 A0A1S3H0H9 A0A2I4D024 A0A3B3QPX9 A0A2I0M5E6 A0A218UIA2 A0A0M8ZST8 A0A0C9R4C0 A0A1L8HN20 A0A3Q3VQI1 A0A3B4WUV3 A0A091QZ03 A0A151NEJ8 K7F2Q5 A0A151NE03 A0A3Q3LKD3 A0A0R4IJ07 A0A0L7RCB5 K7F2Q8 A0A3Q0G9N0 A0A1A7W7B7 A0A3B3CMQ8 I3M7Y6 A0A2U9C4V4 A0A3P9BHT6 V4A9R2 A0A3Q2X553 A0A3P8PZ67 A0A1W5AQ41 A0A3B3YXS1 A0A3Q2X3Y5 A0A087Y518 A0A3Q4HG62 K1Q1G2 A0A3B4F1K3 A0A3Q3IJB3 I3IZA3 A0A3P8W0K3 A0A3Q3F7Y9 A0A3P8VXP7 A0A2A3EJL4 A0A087ZQT9 A0A3P9NNM2 A0A452G121 A0A3P8RRC5 A0A310SHL8 A0A1A8J9B7 A0A3P9L654 A0A154PLA7 M4AXK8 A0A146X6E4 A0A1A8AKL9 A0A1A8VC89 W5PB34 A0A3P9NNT6 G3PX47 U3JES9 H2M688 A0A096MAM9 A0A250YM53 A0A060W5N3 A0A1A8R1T5 A0A1A8R7B7 A0A1A8M2M4 A0A1A8LYF9 A0A1S3FQ83 A0A2B4T0H9 H3D601 A0A146NQN2 A0A1A8FKJ7 A0A452UL10 A0A384DH79 A0A2I2U2Q5 A0A3Q7VLE7 Q8WND5 B2RY98 A0A2K5ECS6 A0A182G4H9 A0A1A8GIZ6 A0A1S4EXL4 A0A2K5J1X1 A0A0K8RU22 Q17MG4 F1Q372 H0V6K7 L5LEY5
PDB
5CQR
E-value=1.24976e-89,
Score=846
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
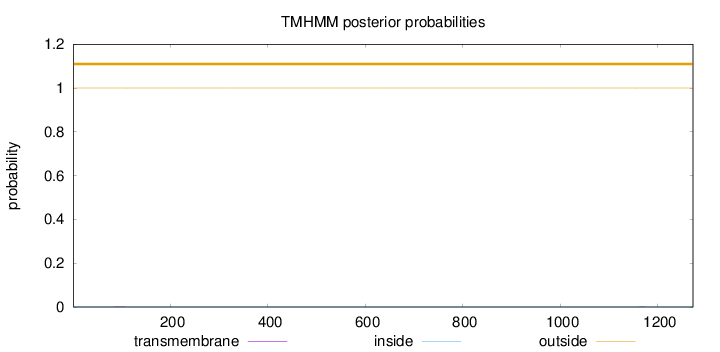
Length:
1272
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01162
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00031
outside
1 - 1272
Population Genetic Test Statistics
Pi
156.261001
Theta
181.992322
Tajima's D
-0.642235
CLR
1.08483
CSRT
0.209489525523724
Interpretation
Uncertain
