Gene
KWMTBOMO12572
Pre Gene Modal
BGIBMGA001530
Annotation
PREDICTED:_small_conductance_calcium-activated_potassium_channel_protein_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.953
Sequence
CDS
ATGGCGGAGAATTTTAGACGAGCATACGGCCCACCTGATGGTGAGTGGTTACCGTCGCCCATGGACTTCAGCAATGCCAGAGGCAGGGCCAAGCCGCTGCCTACCGCTGCAATTGCAGCCTATCGAATTGCAAACGGATTAAGCTTAGGATCCGCATTGACAGAATACTTCGGTGTATTTCAGCTGCGAAAGCCGGTGTCGACACTCTCGATCCCGGGTGCGATGAAGGCTTACTCCGGCGGGGGGCGGGACGCGAGCGGGGAGGAGGCAGGCGTAGCTCTAGTTGGCGTTCACAGCGAGTATCCACGCTTCGGAGAAGAACGGGCTCTGGGCGGCGGCACGTACAAAGGCGGAGGGACAGCCAAGCACAAACCCAACATAGGATATAGCTGGAAGTTTCGATTGAGATGTCTATTAGACGAAAAATCTTTGTTAGAGGTAGCGTCAAGTGGCGTTATAGAATTAAAACAAAAAACTGCTGATGCGGCTGCAAAAGCCTTAATAGTAAAATGTGTACCCGATAAGTACTTGGACATCATAAAAGATGCAACAACTAGCTACACAATGTTAGCCAGTCTGGATAAAGTTTTTGAGAGAAAAGGTATTTTCACTAAATTACACTTGCGAAGAAAGTTATTGTCTCTTAAGCTGAGGGATGATAAATTGGAAGACTATTTCATGAGATTTGAAAGTCTGATTAGAGAAATAGAAAATATTGATAAAAAGATGGATGAAGAAGATAAAATATGTTATTTGCTGACGGGTATGTCGGAAAAGTACAACACAGTGATAACTGCTATTGAAACAATGTCTGCAGATAAAATTGACATGGAGTTTGTCAAGGCTCGGTTATTGGACGAAGAACTCAAACTGAATAATAAAGGTGAAGAGTATAATGATACAAAGAATGATGTAAGTTTTACTGCCTCAAGTGTAGAATGTTATCGTTGCCACAAAAAGGGCCATTTTGCACGTGAATGTAGAAGCAGAGGTGGCTATTCAAGGAGAAACATTAATAATAAAGGTATATGTGGTAAAAACAGAACTGGAGAACAAAATAATATGACAGAAAGCATCCCAGAGAATAAATATAATTTTCTAGCAATAACGGAGTGTAATCTAAGTAGAGAAGACAAATTTATAGAATTTATAATAGATTCAGGAGCTACAGAAAATCTTATCAAGGAAGAATTAGAGAAATACATGTATGAAATTAAAAAACTACCAGATAAAATAAAAATTAAGATTGCAAATGGAGAAATATTAGAGGCTGACAAGAAAGGAAAGATAACCATAAGAGATAAAAATAATAATACTATAAATATAGAAGCGTTAATGGTCCCAGGGATTTATCATAATATAATATCTGTAAGAAACTTAAATATGAAAGGAAATAAAGTTGTTTTTACAAAATTTGGAGCACAAATAATAAATGGAAGAAAAGTCTTAAAATGTTCAGATAGAAATGGTTTATATATAGTACAAGGCGAAATAATACTAATAGAAGAAGCGAACTCAAGTGTACAGCAAGACAAAAATATTTGGCACCAACGTCTCGGACACTTAAATAGGAAATCATTAAAAATTATGGGTTTACCTGTGTCTGATGAAACATGTGGTCCTTGCATGAAGGGAAAATCTACTCGTTTACCATTCTATCCAGTTGACAGGCCAAGAAGTAGAAAAATAGGAGAATTATTGCATACAGATATTGGAGGTCCTGTATCACCAATTTCTTATGATGAAAAACAATATTACCAGACCATAACTGATGATTTTTCTCATTTTGTGACTGTTTATCTATTAAAGAATAAATCTGAAGCCACAAGAAATTTAATAAATCATATAAAGGAGTTAGAAAGACAAAAGGAAACTAAAGTTAAACGCATAAGATGTGATAATGGTGGAGAGTTCACCGCTGGCTGTTTGAAGCAATTTTGTATGGAAAATGGAATTATAATAGAGTATAGTCAACCTTACACACCACAACAAAATGGTGTGGCAGAGAGGCTAAACAGAACATTACTAGATAAGTCAAGAACTTTATTAAGTGAAACTGGATTACCTAAATATTTGTGGTCTGAGGCTATAATGTGTGCAGCATATCAATTGAATCGATGCCCATCAAGATCTATAAAGTATAAGATACCAGCTGAAATATACTACGGACTAGAAAATGTTAACTTGAACAAGTTAAAGGTATTTGGTTCTAAAGCATGGTATACGTGTGTCCCACAGAATAAGAAATTAGATAATAGAGCTGAAGAAGCCATTATGGTTGGCTATGGCAAACAAGGTTATAGATTATGGAATCCAAAAACAACAGAAATAATATTCTGCAGAGATGTCAAGTTTGATGAAAAGTACTATGAACATAAAAACAGTATTAAAAATAACAACTATATTTATTTACCAGAAGAGGACACAAAAAATGAAGTTATGTGCAATGATAAAAATAAAATTGAGGAAGAAAAAGTAATACAGAAAAATGAAGAGACAGACACAAAAAGAACTAGATCAGGAAGGATAATAAAGCAACCTGATTATCTAAAGAATTTTGGGACGTACACAGCATATTGTTACTTAGTGTGTAATGACCCAGAAAGCTATGAAGAAGCGATAAAAGAAGTTTCATGGAAAGAAGCGATACATAAGGAATTAGCAGCACTTGAGAACTTACAAAGCTGGGAAGAGACTAAACTACCGAAAGGAAAATTAGCTATAGACACTAAGTGGGTTTTTAGAACGAAAGACGATGGCACTAAGAAAGCCAGATTAGTCGCTCGAGGTTTTCAAATAAAAAATTATGAGAATGAAATATTCTACAGTCCTGTAGTAAGAATGTCTACGATAAGAGCATTTTTTGTAAAAGCTATCCAAGAAGGATACAAATTAAGGCAATTGGATATTCCTACAGCCTTTCTCAATGGATTTCTTGATAGTGAAGTTTATATAAAAATACCCAAAGGCAAAGAAAACAAAGATGGAATAGTTTTGAGGTTAAAGAGAGCATTGTATGGATTAAAAGAAAGCCCTAAATGCTGGAACATGAGATTTGATGAATTTGCAAGAAAGAATGGATTGATACAGATACAGAAAAAAATTATTAATAAATTGAAGCAAGAATTTAAAGCTAAAGATTTAGGACAGATTGAGAAATATCTTGGTATGGAAATTAAACAGGAAGAAAGTAAATTAACGATTAAACAAGAAAAGATAATTGAGAAGATACTAGAAAAATTTGGAATGTCAGAATGTAAAGGTATAGGAACACCTATGGAGGTAAATTTTCAAATAAACAAAAATGAGAAGATTATTGATGTACCATATCGACAACTCATAGGAAGCATAATGTATTTGGCTACTATGTCCAGACCAGATCTGCAGTTTGCTGTATCATATCTTAGCAGATACCTAGACTGTCCTACTCAACAGACTTGGAAAGCTGGAAAGAGAATTCTGCAATACCTAAAAGCTACAAAAGACAAAATGTTAGTTTTTCAAAAAAGGAATAATCTAGATGTGGATTTATGTGCATATTCAGACTCTGATTGGGCAGGTGATATTGTTGATCGCAAAAGTGTCAGTGGGGGTATCATAATGTGTAGTGATAATCCTGTATTTTGGTACTCTAGAAAACAAAGTTGTGTTGCATTATCGACAGCAGAATCAGAGTACATTGCTAGTGCTTGTTGTGCTCAAGAGTTAGTGTATTTGAAAGGTGTACTGTGTGATTTTGGATATGAATGTAAATTGACTTTATTTATGGATAATAAGGGAGCAATATGTTTAGCGAAGTCAAATGAAAACTCTAAAAGGAGTAAGCATATTGATATAAGGATGCACTATATTAAGGACTTGGTGAGGAATAATGTATTAGATATTGAATATGTGTCATCTGATAAAAATATTGCTGATATTTGTACAAAATCTTTATGCAAAGAAAAGTTAGGTAGGAGAAAGGCGTTATTTGAAAAAAGAAAACGGATAAGTGACTACGCTTTAGTAATGGGGATGTTTGGAATTATAATTATGGTAATAGAAAATGAACTCTCAAGTGCCGGTGTTTATACTAAGGCATCAATATATTCACAGGCGTTAAAGACGCTAATATCAATGTCGACTGTGATTTTACTTGGCCTAATCGTCGCGTATCATGCTTTAGAAGTACAATTATTTATGATAGACAACTGCGCCGACGACTGGCGGATAGCAATGACGTGGCAAAGGATAGCGCAAATCGGCCTCGAACTGGCGATATGCGCCGTTCACCCCATCCCTGGACAATATACTTTTACGTGGACGACAAAGTTGGCCAACAAGGGTGGAGTGATAGGCGTTGAAGTTGTTCCTTATGACGTCACGTTGTCACTGCCGATGTTCTTTAGGCTGTATCTTATATGCAGGGTCATGTTACTTCACAGTAAATTGTTCACGGACGCTTCATCCAGAAGTATAGGAGCTTTAAACAGAATTAATTTTAATACTAGATTCGTCTTGAAGACATTAATGACAATTTGCCCTGGTACTGTATTATTAGTATTTATGGTGTCACTTTGGATAATCGCAAGCTGGACATTACGACAATGCGAAAGGTTTCACGACGAGGAACACGCTAATCTACTGAATGCGATGTGGCTAATAGCTATAACGTTTCTCTCCGTTGGTTTTGGTGACATAGTGCCAAACACATATTGTGGCAGAGGTATTGCTGTTAGCACCGGTATTATGGGAGCTGGATGCACAGCGTTATTAGTCGCGGTTGTTTCAAGAAAATTAGAATTAACTAGAGCCGAGAAGCACGTTCATAACTTCATGATGGACACACAACTTACTAAAAGACTGAAAAACGCAGCGGCTAACGTGCTTCGAGAAACGTGGTTAATATACAAACACACGCGACTAGTAAAAAGAGTTCATCCAGGAAGAGTGAGGACGCATCAGAGGAAGTTTTTGCTAGCTATTTATGCATTACGAAAAGTGAAGATGGATCAACGAAAACTAATGGACAATGCCAACACAATAACTGACATGGCAAAGACACAGAACACGGTCTATGAGATTGTCTCAGATATGAGCACGAGACAGGACACTATAGAAGAGAGGCTGACTAGTTTAGAAGAAAAACTAACAACGTTACAGGAACAAGTAAATAGTTTACCAGATGTTATGGCAAGGTGTTTACAACAATGCTGGGAGAGAGCTGAACAAAGAAGGAATTTTCTTCATCCAGACACCGCCGCTGTGCCTACGCCGCCATCGTCTACTATTACGCCTTATGCAAGAGCAATGCCGTCAGCGTCACATCCGTGGCCCCCGAGTCCGGTGCTGCAACCGTCAGCACGAACCCACTCGGCTCCAGAGAGCACGACAGCTCACTCTCCAACACCAGTCCAGCTGCAGCCTTCAAGCCCTGCTCCTCAAGTGACGCGGCCGCCCTTGCCCAGCTGA
Protein
MAENFRRAYGPPDGEWLPSPMDFSNARGRAKPLPTAAIAAYRIANGLSLGSALTEYFGVFQLRKPVSTLSIPGAMKAYSGGGRDASGEEAGVALVGVHSEYPRFGEERALGGGTYKGGGTAKHKPNIGYSWKFRLRCLLDEKSLLEVASSGVIELKQKTADAAAKALIVKCVPDKYLDIIKDATTSYTMLASLDKVFERKGIFTKLHLRRKLLSLKLRDDKLEDYFMRFESLIREIENIDKKMDEEDKICYLLTGMSEKYNTVITAIETMSADKIDMEFVKARLLDEELKLNNKGEEYNDTKNDVSFTASSVECYRCHKKGHFARECRSRGGYSRRNINNKGICGKNRTGEQNNMTESIPENKYNFLAITECNLSREDKFIEFIIDSGATENLIKEELEKYMYEIKKLPDKIKIKIANGEILEADKKGKITIRDKNNNTINIEALMVPGIYHNIISVRNLNMKGNKVVFTKFGAQIINGRKVLKCSDRNGLYIVQGEIILIEEANSSVQQDKNIWHQRLGHLNRKSLKIMGLPVSDETCGPCMKGKSTRLPFYPVDRPRSRKIGELLHTDIGGPVSPISYDEKQYYQTITDDFSHFVTVYLLKNKSEATRNLINHIKELERQKETKVKRIRCDNGGEFTAGCLKQFCMENGIIIEYSQPYTPQQNGVAERLNRTLLDKSRTLLSETGLPKYLWSEAIMCAAYQLNRCPSRSIKYKIPAEIYYGLENVNLNKLKVFGSKAWYTCVPQNKKLDNRAEEAIMVGYGKQGYRLWNPKTTEIIFCRDVKFDEKYYEHKNSIKNNNYIYLPEEDTKNEVMCNDKNKIEEEKVIQKNEETDTKRTRSGRIIKQPDYLKNFGTYTAYCYLVCNDPESYEEAIKEVSWKEAIHKELAALENLQSWEETKLPKGKLAIDTKWVFRTKDDGTKKARLVARGFQIKNYENEIFYSPVVRMSTIRAFFVKAIQEGYKLRQLDIPTAFLNGFLDSEVYIKIPKGKENKDGIVLRLKRALYGLKESPKCWNMRFDEFARKNGLIQIQKKIINKLKQEFKAKDLGQIEKYLGMEIKQEESKLTIKQEKIIEKILEKFGMSECKGIGTPMEVNFQINKNEKIIDVPYRQLIGSIMYLATMSRPDLQFAVSYLSRYLDCPTQQTWKAGKRILQYLKATKDKMLVFQKRNNLDVDLCAYSDSDWAGDIVDRKSVSGGIIMCSDNPVFWYSRKQSCVALSTAESEYIASACCAQELVYLKGVLCDFGYECKLTLFMDNKGAICLAKSNENSKRSKHIDIRMHYIKDLVRNNVLDIEYVSSDKNIADICTKSLCKEKLGRRKALFEKRKRISDYALVMGMFGIIIMVIENELSSAGVYTKASIYSQALKTLISMSTVILLGLIVAYHALEVQLFMIDNCADDWRIAMTWQRIAQIGLELAICAVHPIPGQYTFTWTTKLANKGGVIGVEVVPYDVTLSLPMFFRLYLICRVMLLHSKLFTDASSRSIGALNRINFNTRFVLKTLMTICPGTVLLVFMVSLWIIASWTLRQCERFHDEEHANLLNAMWLIAITFLSVGFGDIVPNTYCGRGIAVSTGIMGAGCTALLVAVVSRKLELTRAEKHVHNFMMDTQLTKRLKNAAANVLRETWLIYKHTRLVKRVHPGRVRTHQRKFLLAIYALRKVKMDQRKLMDNANTITDMAKTQNTVYEIVSDMSTRQDTIEERLTSLEEKLTTLQEQVNSLPDVMARCLQQCWERAEQRRNFLHPDTAAVPTPPSSTITPYARAMPSASHPWPPSPVLQPSARTHSAPESTTAHSPTPVQLQPSSPAPQVTRPPLPS
Summary
Uniprot
Interpro
Gene 3D
ProteinModelPortal
PDB
6CNO
E-value=7.89115e-75,
Score=720
Ontologies
GO
PANTHER
Topology
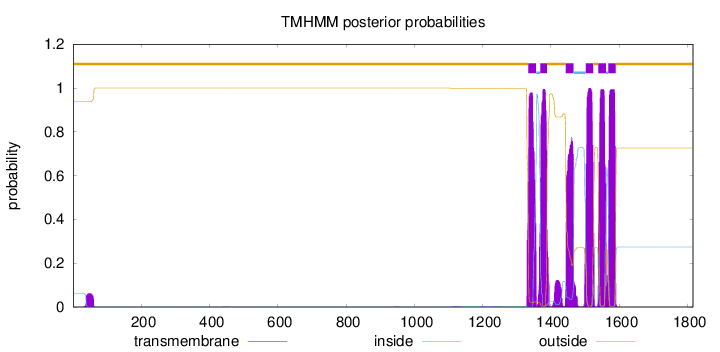
Length:
1816
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
128.39194
Exp number, first 60 AAs:
1.36523
Total prob of N-in:
0.06230
outside
1 - 1333
TMhelix
1334 - 1356
inside
1357 - 1368
TMhelix
1369 - 1388
outside
1389 - 1443
TMhelix
1444 - 1466
inside
1467 - 1501
TMhelix
1502 - 1524
outside
1525 - 1538
TMhelix
1539 - 1561
inside
1562 - 1567
TMhelix
1568 - 1590
outside
1591 - 1816
Population Genetic Test Statistics
Pi
187.34297
Theta
185.848767
Tajima's D
0.304123
CLR
0.499166
CSRT
0.462776861156942
Interpretation
Uncertain
