Pre Gene Modal
BGIBMGA001448
Annotation
PREDICTED:_arginine-glutamic_acid_dipeptide_repeats_protein-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.664
Sequence
CDS
ATGCTAAAGAAACGCCAAGAGGCCGAACGTCTTGCTGCCGCCGGAAAACACAAGTACGAGTACGACAGCGACGAGGACACAACAGACGGTACCTGGGAACATAAATTGCGTGCTAAAGAAATGTTTGCTACGGAGAAGTGGGCCGATGAATTAACACGGCAGGCTGCTGGTAAACATCACATTGGAGATTTCCTGCCTCCGGAGGAATTGAAAAAATTCATGGAAAAATATTCAGCTTTGAAAAATGGCAAAGAGCCAGACCTGAGTGATTATAAAGAGTTCAAGCTGAAGGAAGATAATGTCGGGTTTAAAATGCTTCAGAAGCTTGGATGGAATGAGGGTCAAGGTCTCGGCGTAGAAGGTTCAGGCATCGTGGAACCTATCAACAAGGCGAAACAACCAGTACAGAATCTAGGACTGGGCGCGTCCACTTCGGACGTGGTGTCGCCCGATGACGACGAGTTCGACGCGTACAGGAAACGCATGATGCTCGCTTACAGATTCCGACCTAATCCATTGAATAATCCACGCAGACCATATTATTAA
Protein
MLKKRQEAERLAAAGKHKYEYDSDEDTTDGTWEHKLRAKEMFATEKWADELTRQAAGKHHIGDFLPPEELKKFMEKYSALKNGKEPDLSDYKEFKLKEDNVGFKMLQKLGWNEGQGLGVEGSGIVEPINKAKQPVQNLGLGASTSDVVSPDDDEFDAYRKRMMLAYRFRPNPLNNPRRPYY
Summary
Uniprot
H9IW17
A0A2A4IVW2
A0A1E1WH47
A0A1E1WQN0
A0A3S2LJD9
A0A2H1V0I7
+ More
A0A212F2A5 A0A194Q8D2 A0A0C9R1T8 A0A182M8S8 A0A2J7PE68 A0A2A3EHH0 A0A023EXE5 A0A151ISQ1 A0A195BPP8 A0A026WY64 A0A0C9QL21 A0A151XGJ6 A0A151ID21 A0A0L7QMY5 Q17P48 A0A310SF69 F4WTS5 A0A2J7PE63 A0A158N9N2 A0A3L8D444 E2BRI8 E2A8Q2 A0A087ZU44 A0A0N0BHX7 A0A2P8Y9C2 A0A154PT82 A0A1Q3FGN9 A0A1S4EW60 A0A1Q3FGS8 A0A1Q3FGI0 A0A2S2NUX6 A0A2H8TNK1 A0A182STW4 A0A182PE56 A0A182ME68 A0A182N596 A0A1B6LPC8 A0A182JNU5 A0A182VQR4 A0A182J3W6 A0A1W4UWR9 W8B6D1 A0A182QZY3 A0A2J7PE73 A0A084WSN3 E0VRM1 A0A2J7PE85 A0A1B6DWE6 A0A182YM39 A0A182TZR7 A0A182RIX8 A0A0R1E4W8 A0A1B6GDR8 Q9VNC5 A0A182LCJ1 A0A1S4GHB5 Q7QBC6 A0A1B0DL25 A0A1B6D514 A0A182HJB2 A0A182UQT2 A0A182XE09 A0A0R1E086 A0A1I8MKY2 A0A1B6DSM7 A0A2M4A9F4 A0A2S2QMG3 T1PDB5 W5J9M3 A0A2M4BBN6 A0A2M4CLE5 A0A2M3Z152 A0A2M4A7M2 A0A2M4A7H8 A0A182FSJ8 A0A2M4BBP9 A0A1I8M0S0 A0A1Y1KTS4 A0A1Y1KSE4 B4GF85 I5APC6 A0A1L8DFK1 A0A1A9X1V6 A0A1Y1KSE7 A0A1Y1KW64 A0A1W4UJA1 B3LZ41 B4PVA8 A0A1Y1KNX1 A0A3B0JVD8 Q9VNC4 A0A1Y1KR84 A0A0B4KF34 B4I412
A0A212F2A5 A0A194Q8D2 A0A0C9R1T8 A0A182M8S8 A0A2J7PE68 A0A2A3EHH0 A0A023EXE5 A0A151ISQ1 A0A195BPP8 A0A026WY64 A0A0C9QL21 A0A151XGJ6 A0A151ID21 A0A0L7QMY5 Q17P48 A0A310SF69 F4WTS5 A0A2J7PE63 A0A158N9N2 A0A3L8D444 E2BRI8 E2A8Q2 A0A087ZU44 A0A0N0BHX7 A0A2P8Y9C2 A0A154PT82 A0A1Q3FGN9 A0A1S4EW60 A0A1Q3FGS8 A0A1Q3FGI0 A0A2S2NUX6 A0A2H8TNK1 A0A182STW4 A0A182PE56 A0A182ME68 A0A182N596 A0A1B6LPC8 A0A182JNU5 A0A182VQR4 A0A182J3W6 A0A1W4UWR9 W8B6D1 A0A182QZY3 A0A2J7PE73 A0A084WSN3 E0VRM1 A0A2J7PE85 A0A1B6DWE6 A0A182YM39 A0A182TZR7 A0A182RIX8 A0A0R1E4W8 A0A1B6GDR8 Q9VNC5 A0A182LCJ1 A0A1S4GHB5 Q7QBC6 A0A1B0DL25 A0A1B6D514 A0A182HJB2 A0A182UQT2 A0A182XE09 A0A0R1E086 A0A1I8MKY2 A0A1B6DSM7 A0A2M4A9F4 A0A2S2QMG3 T1PDB5 W5J9M3 A0A2M4BBN6 A0A2M4CLE5 A0A2M3Z152 A0A2M4A7M2 A0A2M4A7H8 A0A182FSJ8 A0A2M4BBP9 A0A1I8M0S0 A0A1Y1KTS4 A0A1Y1KSE4 B4GF85 I5APC6 A0A1L8DFK1 A0A1A9X1V6 A0A1Y1KSE7 A0A1Y1KW64 A0A1W4UJA1 B3LZ41 B4PVA8 A0A1Y1KNX1 A0A3B0JVD8 Q9VNC4 A0A1Y1KR84 A0A0B4KF34 B4I412
Pubmed
EMBL
BABH01006150
BABH01006151
BABH01006152
NWSH01005600
PCG64107.1
GDQN01004766
+ More
JAT86288.1 GDQN01001762 JAT89292.1 RSAL01000092 RVE47955.1 ODYU01000122 SOQ34360.1 AGBW02010772 OWR47875.1 KQ459564 KPI99665.1 GBYB01000811 JAG70578.1 AXCM01008055 NEVH01026108 PNF14630.1 KZ288260 PBC30451.1 GAPW01000504 JAC13094.1 KQ981064 KYN09892.1 KQ976428 KYM87815.1 KK107083 EZA60074.1 GBYB01001242 JAG71009.1 KQ982169 KYQ59524.1 KQ977991 KYM98292.1 KQ414868 KOC59983.1 CH477194 EAT48449.1 KQ769378 OAD52869.1 GL888344 EGI62390.1 PNF14631.1 ADTU01009716 ADTU01009717 QOIP01000014 RLU14994.1 GL449965 EFN81680.1 GL437663 EFN70131.1 KQ435737 KOX76884.1 PYGN01000783 PSN40857.1 KQ435066 KZC14320.1 GFDL01008336 JAV26709.1 GFDL01008333 JAV26712.1 GFDL01008385 JAV26660.1 GGMR01008249 MBY20868.1 GFXV01003942 MBW15747.1 AXCM01001496 GEBQ01014623 JAT25354.1 GAMC01013965 GAMC01013962 JAB92593.1 AXCN02000148 PNF14629.1 ATLV01026629 ATLV01026630 KE525415 KFB53227.1 DS235478 EEB16027.1 PNF14628.1 GEDC01015643 GEDC01007305 JAS21655.1 JAS29993.1 CM000160 KRK02714.1 GECZ01009177 JAS60592.1 AY060628 BT024958 AAL28176.1 ABE01188.1 AAAB01008879 EAA08349.5 AJVK01069052 GEDC01016506 JAS20792.1 APCN01002135 KRK02716.1 GEDC01008614 JAS28684.1 GGFK01004074 MBW37395.1 GGMS01009742 MBY78945.1 KA646110 AFP60739.1 ADMH02002074 ETN59555.1 GGFJ01001315 MBW50456.1 GGFL01002004 MBW66182.1 GGFM01001493 MBW22244.1 GGFK01003434 MBW36755.1 GGFK01003435 MBW36756.1 GGFJ01001316 MBW50457.1 GEZM01077964 JAV63105.1 GEZM01077968 JAV63100.1 CH479182 EDW34270.1 CM000070 EIM52811.2 GFDF01008842 JAV05242.1 GEZM01077966 GEZM01077965 GEZM01077961 JAV63110.1 GEZM01077970 GEZM01077963 GEZM01077959 JAV63107.1 CH902617 EDV41915.2 EDW95777.2 GEZM01077962 GEZM01077960 JAV63109.1 OUUW01000008 SPP84372.1 AE014297 AAF52020.2 GEZM01077969 GEZM01077967 JAV63098.1 AGB95668.1 CH480821 EDW54955.1
JAT86288.1 GDQN01001762 JAT89292.1 RSAL01000092 RVE47955.1 ODYU01000122 SOQ34360.1 AGBW02010772 OWR47875.1 KQ459564 KPI99665.1 GBYB01000811 JAG70578.1 AXCM01008055 NEVH01026108 PNF14630.1 KZ288260 PBC30451.1 GAPW01000504 JAC13094.1 KQ981064 KYN09892.1 KQ976428 KYM87815.1 KK107083 EZA60074.1 GBYB01001242 JAG71009.1 KQ982169 KYQ59524.1 KQ977991 KYM98292.1 KQ414868 KOC59983.1 CH477194 EAT48449.1 KQ769378 OAD52869.1 GL888344 EGI62390.1 PNF14631.1 ADTU01009716 ADTU01009717 QOIP01000014 RLU14994.1 GL449965 EFN81680.1 GL437663 EFN70131.1 KQ435737 KOX76884.1 PYGN01000783 PSN40857.1 KQ435066 KZC14320.1 GFDL01008336 JAV26709.1 GFDL01008333 JAV26712.1 GFDL01008385 JAV26660.1 GGMR01008249 MBY20868.1 GFXV01003942 MBW15747.1 AXCM01001496 GEBQ01014623 JAT25354.1 GAMC01013965 GAMC01013962 JAB92593.1 AXCN02000148 PNF14629.1 ATLV01026629 ATLV01026630 KE525415 KFB53227.1 DS235478 EEB16027.1 PNF14628.1 GEDC01015643 GEDC01007305 JAS21655.1 JAS29993.1 CM000160 KRK02714.1 GECZ01009177 JAS60592.1 AY060628 BT024958 AAL28176.1 ABE01188.1 AAAB01008879 EAA08349.5 AJVK01069052 GEDC01016506 JAS20792.1 APCN01002135 KRK02716.1 GEDC01008614 JAS28684.1 GGFK01004074 MBW37395.1 GGMS01009742 MBY78945.1 KA646110 AFP60739.1 ADMH02002074 ETN59555.1 GGFJ01001315 MBW50456.1 GGFL01002004 MBW66182.1 GGFM01001493 MBW22244.1 GGFK01003434 MBW36755.1 GGFK01003435 MBW36756.1 GGFJ01001316 MBW50457.1 GEZM01077964 JAV63105.1 GEZM01077968 JAV63100.1 CH479182 EDW34270.1 CM000070 EIM52811.2 GFDF01008842 JAV05242.1 GEZM01077966 GEZM01077965 GEZM01077961 JAV63110.1 GEZM01077970 GEZM01077963 GEZM01077959 JAV63107.1 CH902617 EDV41915.2 EDW95777.2 GEZM01077962 GEZM01077960 JAV63109.1 OUUW01000008 SPP84372.1 AE014297 AAF52020.2 GEZM01077969 GEZM01077967 JAV63098.1 AGB95668.1 CH480821 EDW54955.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000075883
+ More
UP000235965 UP000242457 UP000078492 UP000078540 UP000053097 UP000075809 UP000078542 UP000053825 UP000008820 UP000007755 UP000005205 UP000279307 UP000008237 UP000000311 UP000005203 UP000053105 UP000245037 UP000076502 UP000075901 UP000075885 UP000075884 UP000075881 UP000075920 UP000075880 UP000192221 UP000075886 UP000030765 UP000009046 UP000076408 UP000075902 UP000075900 UP000002282 UP000075882 UP000007062 UP000092462 UP000075840 UP000075903 UP000076407 UP000095301 UP000000673 UP000069272 UP000008744 UP000001819 UP000091820 UP000007801 UP000268350 UP000000803 UP000001292
UP000235965 UP000242457 UP000078492 UP000078540 UP000053097 UP000075809 UP000078542 UP000053825 UP000008820 UP000007755 UP000005205 UP000279307 UP000008237 UP000000311 UP000005203 UP000053105 UP000245037 UP000076502 UP000075901 UP000075885 UP000075884 UP000075881 UP000075920 UP000075880 UP000192221 UP000075886 UP000030765 UP000009046 UP000076408 UP000075902 UP000075900 UP000002282 UP000075882 UP000007062 UP000092462 UP000075840 UP000075903 UP000076407 UP000095301 UP000000673 UP000069272 UP000008744 UP000001819 UP000091820 UP000007801 UP000268350 UP000000803 UP000001292
SUPFAM
SSF109905
SSF109905
Gene 3D
ProteinModelPortal
H9IW17
A0A2A4IVW2
A0A1E1WH47
A0A1E1WQN0
A0A3S2LJD9
A0A2H1V0I7
+ More
A0A212F2A5 A0A194Q8D2 A0A0C9R1T8 A0A182M8S8 A0A2J7PE68 A0A2A3EHH0 A0A023EXE5 A0A151ISQ1 A0A195BPP8 A0A026WY64 A0A0C9QL21 A0A151XGJ6 A0A151ID21 A0A0L7QMY5 Q17P48 A0A310SF69 F4WTS5 A0A2J7PE63 A0A158N9N2 A0A3L8D444 E2BRI8 E2A8Q2 A0A087ZU44 A0A0N0BHX7 A0A2P8Y9C2 A0A154PT82 A0A1Q3FGN9 A0A1S4EW60 A0A1Q3FGS8 A0A1Q3FGI0 A0A2S2NUX6 A0A2H8TNK1 A0A182STW4 A0A182PE56 A0A182ME68 A0A182N596 A0A1B6LPC8 A0A182JNU5 A0A182VQR4 A0A182J3W6 A0A1W4UWR9 W8B6D1 A0A182QZY3 A0A2J7PE73 A0A084WSN3 E0VRM1 A0A2J7PE85 A0A1B6DWE6 A0A182YM39 A0A182TZR7 A0A182RIX8 A0A0R1E4W8 A0A1B6GDR8 Q9VNC5 A0A182LCJ1 A0A1S4GHB5 Q7QBC6 A0A1B0DL25 A0A1B6D514 A0A182HJB2 A0A182UQT2 A0A182XE09 A0A0R1E086 A0A1I8MKY2 A0A1B6DSM7 A0A2M4A9F4 A0A2S2QMG3 T1PDB5 W5J9M3 A0A2M4BBN6 A0A2M4CLE5 A0A2M3Z152 A0A2M4A7M2 A0A2M4A7H8 A0A182FSJ8 A0A2M4BBP9 A0A1I8M0S0 A0A1Y1KTS4 A0A1Y1KSE4 B4GF85 I5APC6 A0A1L8DFK1 A0A1A9X1V6 A0A1Y1KSE7 A0A1Y1KW64 A0A1W4UJA1 B3LZ41 B4PVA8 A0A1Y1KNX1 A0A3B0JVD8 Q9VNC4 A0A1Y1KR84 A0A0B4KF34 B4I412
A0A212F2A5 A0A194Q8D2 A0A0C9R1T8 A0A182M8S8 A0A2J7PE68 A0A2A3EHH0 A0A023EXE5 A0A151ISQ1 A0A195BPP8 A0A026WY64 A0A0C9QL21 A0A151XGJ6 A0A151ID21 A0A0L7QMY5 Q17P48 A0A310SF69 F4WTS5 A0A2J7PE63 A0A158N9N2 A0A3L8D444 E2BRI8 E2A8Q2 A0A087ZU44 A0A0N0BHX7 A0A2P8Y9C2 A0A154PT82 A0A1Q3FGN9 A0A1S4EW60 A0A1Q3FGS8 A0A1Q3FGI0 A0A2S2NUX6 A0A2H8TNK1 A0A182STW4 A0A182PE56 A0A182ME68 A0A182N596 A0A1B6LPC8 A0A182JNU5 A0A182VQR4 A0A182J3W6 A0A1W4UWR9 W8B6D1 A0A182QZY3 A0A2J7PE73 A0A084WSN3 E0VRM1 A0A2J7PE85 A0A1B6DWE6 A0A182YM39 A0A182TZR7 A0A182RIX8 A0A0R1E4W8 A0A1B6GDR8 Q9VNC5 A0A182LCJ1 A0A1S4GHB5 Q7QBC6 A0A1B0DL25 A0A1B6D514 A0A182HJB2 A0A182UQT2 A0A182XE09 A0A0R1E086 A0A1I8MKY2 A0A1B6DSM7 A0A2M4A9F4 A0A2S2QMG3 T1PDB5 W5J9M3 A0A2M4BBN6 A0A2M4CLE5 A0A2M3Z152 A0A2M4A7M2 A0A2M4A7H8 A0A182FSJ8 A0A2M4BBP9 A0A1I8M0S0 A0A1Y1KTS4 A0A1Y1KSE4 B4GF85 I5APC6 A0A1L8DFK1 A0A1A9X1V6 A0A1Y1KSE7 A0A1Y1KW64 A0A1W4UJA1 B3LZ41 B4PVA8 A0A1Y1KNX1 A0A3B0JVD8 Q9VNC4 A0A1Y1KR84 A0A0B4KF34 B4I412
PDB
5Y88
E-value=0.000256527,
Score=101
Ontologies
GO
PANTHER
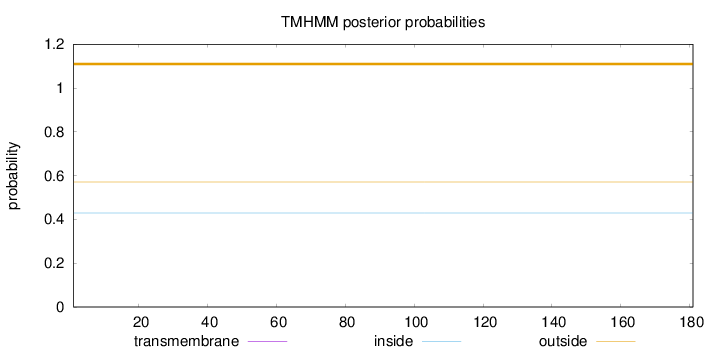
Topology
Length:
181
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.42929
outside
1 - 181
Population Genetic Test Statistics
Pi
247.330163
Theta
192.023341
Tajima's D
1.049309
CLR
0.001931
CSRT
0.67821608919554
Interpretation
Uncertain
