Gene
KWMTBOMO12541
Pre Gene Modal
BGIBMGA001545
Annotation
PREDICTED:_hemicentin-2-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.353
Sequence
CDS
ATGTTCATTTTGAAGGCTGTCAAAGCAAATACTCCTGAAGCAAACAATATAGGTTTGGTGTCGTGGATTCGGAAACGGGATTTGCACATACTAACCGTCGGCGTGCACACGTATAGCAGCGACGCGAGATTCGCGGCCCTACACACGGACGGGTCAGACGAATGGACTTTACGCGTCGCCCCAGCGCAGCCGAGAGACTCCGGCTCGTATGAATGTCAAGTGTCCACTGAACCGAAAATAAGCCTTTCTTTTAGGCTCACAGTTGTCGTATCAAAGGCAGAAATACTGTCGGGACCAGAACTCTTTGTGCGAGCCGGGTCCGACATCAATCTGACATGTATCGCTAGACATGCCCCAGATCCGCCAAGGTAA
Protein
MFILKAVKANTPEANNIGLVSWIRKRDLHILTVGVHTYSSDARFAALHTDGSDEWTLRVAPAQPRDSGSYECQVSTEPKISLSFRLTVVVSKAEILSGPELFVRAGSDINLTCIARHAPDPPR
Summary
Uniprot
A0A2H1VFR0
A0A2A4JRH3
H9IWB4
A0A212ELK7
A0A3S2LUX2
A0A2H1WDR3
+ More
A0A2A4JM26 A0A212F480 A0A0N1IP48 A0A194Q4T7 A0A1B0FNB0 A0A1B0AJ69 A0A182YHY9 A0A1A9UWE0 A0A1A9WJ22 A0A0L0C3D7 A0A1I8MR61 A0A1I8QBZ1 W8BEW6 A0A1Y9J0M5 A0A1S4GBG5 A0A336KD98 Q7PX67 A0A182QLK4 A0A1Y9IVC5 A0A2P8YWI1 A0A1J1IMB4 A0A1Y9IS36 A0A182IYA3 Q179I1 A0A2J7PVA2 A0A182H4P8 A0A084W049 A0A182GLY2 A0A182YQ25 A0A182T454 A0A182J1H6 W5J598 A0A182FS04 A0A3Q0JLL4 A0A182TGW3 B0WFL2 A0A1B6MCG7 D6WKJ3 Q7PUC4 A0A182RA57 A0A182NQP9 A0A182KAT0 Q294Z8 A0A182PRS9 B4KDZ6 A0A3B0KP62 B4NAW4 Q5U198 B4JUP6 Q59DX6 B4M5D4 B3P4J7 A0A1W4WDJ3 A0A0M4EP47 B4PM67 B4QU29 A0A182HKE1 A0A182X9F5 A0A1J1IH72 B4HI17 A0A1S4GBB1 A0A182VFY5 A0A1Y9IVK1 A0A224XKD3 T1I4D7 A0A182MCL2 A0A182QP59 A0A2S2R2Z7 J9L9Q8 A0A2H8U2T4 A0A088ARL1 B3M0X9 A0A084W055 B4GME3 A0A0C9PJB4 A0A195FFZ1 A0A195DUW5 E2AIN0 A0A151XIL3 A0A026WXN1 A0A0K8W5Q0 K7J0F9 A0A1I8NCT4 A0A034WNE9 F4WN38 A0A2A3E2L8 A0A0M3QXG1 B4M5D3 B4JUP5 E2C4Z5 A0A0A1WHJ4 W8BV78 B4KDZ5 A0A1I8PJB7
A0A2A4JM26 A0A212F480 A0A0N1IP48 A0A194Q4T7 A0A1B0FNB0 A0A1B0AJ69 A0A182YHY9 A0A1A9UWE0 A0A1A9WJ22 A0A0L0C3D7 A0A1I8MR61 A0A1I8QBZ1 W8BEW6 A0A1Y9J0M5 A0A1S4GBG5 A0A336KD98 Q7PX67 A0A182QLK4 A0A1Y9IVC5 A0A2P8YWI1 A0A1J1IMB4 A0A1Y9IS36 A0A182IYA3 Q179I1 A0A2J7PVA2 A0A182H4P8 A0A084W049 A0A182GLY2 A0A182YQ25 A0A182T454 A0A182J1H6 W5J598 A0A182FS04 A0A3Q0JLL4 A0A182TGW3 B0WFL2 A0A1B6MCG7 D6WKJ3 Q7PUC4 A0A182RA57 A0A182NQP9 A0A182KAT0 Q294Z8 A0A182PRS9 B4KDZ6 A0A3B0KP62 B4NAW4 Q5U198 B4JUP6 Q59DX6 B4M5D4 B3P4J7 A0A1W4WDJ3 A0A0M4EP47 B4PM67 B4QU29 A0A182HKE1 A0A182X9F5 A0A1J1IH72 B4HI17 A0A1S4GBB1 A0A182VFY5 A0A1Y9IVK1 A0A224XKD3 T1I4D7 A0A182MCL2 A0A182QP59 A0A2S2R2Z7 J9L9Q8 A0A2H8U2T4 A0A088ARL1 B3M0X9 A0A084W055 B4GME3 A0A0C9PJB4 A0A195FFZ1 A0A195DUW5 E2AIN0 A0A151XIL3 A0A026WXN1 A0A0K8W5Q0 K7J0F9 A0A1I8NCT4 A0A034WNE9 F4WN38 A0A2A3E2L8 A0A0M3QXG1 B4M5D3 B4JUP5 E2C4Z5 A0A0A1WHJ4 W8BV78 B4KDZ5 A0A1I8PJB7
Pubmed
19121390
22118469
26354079
25244985
26108605
25315136
+ More
24495485 12364791 29403074 17510324 26483478 24438588 20920257 23761445 18362917 19820115 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30467080 17550304 20798317 24508170 20075255 25348373 21719571 25830018
24495485 12364791 29403074 17510324 26483478 24438588 20920257 23761445 18362917 19820115 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30467080 17550304 20798317 24508170 20075255 25348373 21719571 25830018
EMBL
ODYU01002335
SOQ39700.1
NWSH01000717
PCG74635.1
BABH01006318
AGBW02014033
+ More
OWR42374.1 RSAL01000006 RVE54304.1 ODYU01007970 SOQ51191.1 NWSH01001012 PCG73135.1 AGBW02010425 OWR48539.1 KQ460622 KPJ13511.1 KQ459585 KPI98420.1 CCAG010016377 JRES01000953 KNC26850.1 GAMC01006680 JAB99875.1 AAAB01008987 UFQS01000083 UFQT01000083 SSW99060.1 SSX19442.1 EAA01782.5 AXCN02001506 AXCN02001507 AXCN02001508 PYGN01000321 PSN48575.1 CVRI01000051 CRK99609.1 CH477349 EAT42909.1 NEVH01020980 PNF20251.1 JXUM01109976 JXUM01109977 KQ565370 KXJ71043.1 ATLV01019088 ATLV01019089 KE525262 KFB43593.1 JXUM01073826 JXUM01073827 JXUM01073828 KQ562797 KXJ75108.1 ADMH02002101 ETN59156.1 DS231918 EDS26280.1 GEBQ01006398 JAT33579.1 KQ971342 EFA03572.1 EAA01771.6 CM000070 EAL28814.2 CH933806 EDW16017.1 OUUW01000014 SPP88409.1 CH964232 EDW80928.2 BT015994 AAV36879.1 CH916374 EDV91216.1 AE014297 BT057996 AAX52946.2 ACM16706.1 CH940652 EDW59845.1 CH954181 EDV49512.1 CP012526 ALC45810.1 CM000160 EDW96933.1 CM000364 EDX13360.1 APCN01000826 CRK99597.1 CH480815 EDW42599.1 GFTR01003489 JAW12937.1 ACPB03003817 ACPB03003818 ACPB03003819 AXCM01000701 GGMS01015142 MBY84345.1 ABLF02039845 ABLF02039849 ABLF02039858 ABLF02039860 GFXV01008073 MBW19878.1 CH902617 EDV43208.2 ATLV01019091 KFB43599.1 CH479185 EDW38017.1 GBYB01001053 JAG70820.1 KQ981617 KYN39276.1 KQ980322 KYN16668.1 GL439840 EFN66719.1 KQ982080 KYQ60128.1 KK107064 EZA60820.1 GDHF01005940 JAI46374.1 GAKP01001851 JAC57101.1 GL888234 EGI64380.1 KZ288457 PBC25506.1 ALC45809.1 EDW59844.2 EDV91215.1 GL452712 EFN76976.1 GBXI01016449 JAC97842.1 GAMC01009344 GAMC01009343 JAB97211.1 EDW16016.1
OWR42374.1 RSAL01000006 RVE54304.1 ODYU01007970 SOQ51191.1 NWSH01001012 PCG73135.1 AGBW02010425 OWR48539.1 KQ460622 KPJ13511.1 KQ459585 KPI98420.1 CCAG010016377 JRES01000953 KNC26850.1 GAMC01006680 JAB99875.1 AAAB01008987 UFQS01000083 UFQT01000083 SSW99060.1 SSX19442.1 EAA01782.5 AXCN02001506 AXCN02001507 AXCN02001508 PYGN01000321 PSN48575.1 CVRI01000051 CRK99609.1 CH477349 EAT42909.1 NEVH01020980 PNF20251.1 JXUM01109976 JXUM01109977 KQ565370 KXJ71043.1 ATLV01019088 ATLV01019089 KE525262 KFB43593.1 JXUM01073826 JXUM01073827 JXUM01073828 KQ562797 KXJ75108.1 ADMH02002101 ETN59156.1 DS231918 EDS26280.1 GEBQ01006398 JAT33579.1 KQ971342 EFA03572.1 EAA01771.6 CM000070 EAL28814.2 CH933806 EDW16017.1 OUUW01000014 SPP88409.1 CH964232 EDW80928.2 BT015994 AAV36879.1 CH916374 EDV91216.1 AE014297 BT057996 AAX52946.2 ACM16706.1 CH940652 EDW59845.1 CH954181 EDV49512.1 CP012526 ALC45810.1 CM000160 EDW96933.1 CM000364 EDX13360.1 APCN01000826 CRK99597.1 CH480815 EDW42599.1 GFTR01003489 JAW12937.1 ACPB03003817 ACPB03003818 ACPB03003819 AXCM01000701 GGMS01015142 MBY84345.1 ABLF02039845 ABLF02039849 ABLF02039858 ABLF02039860 GFXV01008073 MBW19878.1 CH902617 EDV43208.2 ATLV01019091 KFB43599.1 CH479185 EDW38017.1 GBYB01001053 JAG70820.1 KQ981617 KYN39276.1 KQ980322 KYN16668.1 GL439840 EFN66719.1 KQ982080 KYQ60128.1 KK107064 EZA60820.1 GDHF01005940 JAI46374.1 GAKP01001851 JAC57101.1 GL888234 EGI64380.1 KZ288457 PBC25506.1 ALC45809.1 EDW59844.2 EDV91215.1 GL452712 EFN76976.1 GBXI01016449 JAC97842.1 GAMC01009344 GAMC01009343 JAB97211.1 EDW16016.1
Proteomes
UP000218220
UP000005204
UP000007151
UP000283053
UP000053240
UP000053268
+ More
UP000092444 UP000092445 UP000076408 UP000078200 UP000091820 UP000037069 UP000095301 UP000095300 UP000076407 UP000007062 UP000075886 UP000075920 UP000245037 UP000183832 UP000075903 UP000075880 UP000008820 UP000235965 UP000069940 UP000249989 UP000030765 UP000075901 UP000000673 UP000069272 UP000079169 UP000075902 UP000002320 UP000007266 UP000075900 UP000075884 UP000075881 UP000001819 UP000075885 UP000009192 UP000268350 UP000007798 UP000001070 UP000000803 UP000008792 UP000008711 UP000192221 UP000092553 UP000002282 UP000000304 UP000075840 UP000001292 UP000015103 UP000075883 UP000007819 UP000005203 UP000007801 UP000008744 UP000078541 UP000078492 UP000000311 UP000075809 UP000053097 UP000002358 UP000007755 UP000242457 UP000008237
UP000092444 UP000092445 UP000076408 UP000078200 UP000091820 UP000037069 UP000095301 UP000095300 UP000076407 UP000007062 UP000075886 UP000075920 UP000245037 UP000183832 UP000075903 UP000075880 UP000008820 UP000235965 UP000069940 UP000249989 UP000030765 UP000075901 UP000000673 UP000069272 UP000079169 UP000075902 UP000002320 UP000007266 UP000075900 UP000075884 UP000075881 UP000001819 UP000075885 UP000009192 UP000268350 UP000007798 UP000001070 UP000000803 UP000008792 UP000008711 UP000192221 UP000092553 UP000002282 UP000000304 UP000075840 UP000001292 UP000015103 UP000075883 UP000007819 UP000005203 UP000007801 UP000008744 UP000078541 UP000078492 UP000000311 UP000075809 UP000053097 UP000002358 UP000007755 UP000242457 UP000008237
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A2H1VFR0
A0A2A4JRH3
H9IWB4
A0A212ELK7
A0A3S2LUX2
A0A2H1WDR3
+ More
A0A2A4JM26 A0A212F480 A0A0N1IP48 A0A194Q4T7 A0A1B0FNB0 A0A1B0AJ69 A0A182YHY9 A0A1A9UWE0 A0A1A9WJ22 A0A0L0C3D7 A0A1I8MR61 A0A1I8QBZ1 W8BEW6 A0A1Y9J0M5 A0A1S4GBG5 A0A336KD98 Q7PX67 A0A182QLK4 A0A1Y9IVC5 A0A2P8YWI1 A0A1J1IMB4 A0A1Y9IS36 A0A182IYA3 Q179I1 A0A2J7PVA2 A0A182H4P8 A0A084W049 A0A182GLY2 A0A182YQ25 A0A182T454 A0A182J1H6 W5J598 A0A182FS04 A0A3Q0JLL4 A0A182TGW3 B0WFL2 A0A1B6MCG7 D6WKJ3 Q7PUC4 A0A182RA57 A0A182NQP9 A0A182KAT0 Q294Z8 A0A182PRS9 B4KDZ6 A0A3B0KP62 B4NAW4 Q5U198 B4JUP6 Q59DX6 B4M5D4 B3P4J7 A0A1W4WDJ3 A0A0M4EP47 B4PM67 B4QU29 A0A182HKE1 A0A182X9F5 A0A1J1IH72 B4HI17 A0A1S4GBB1 A0A182VFY5 A0A1Y9IVK1 A0A224XKD3 T1I4D7 A0A182MCL2 A0A182QP59 A0A2S2R2Z7 J9L9Q8 A0A2H8U2T4 A0A088ARL1 B3M0X9 A0A084W055 B4GME3 A0A0C9PJB4 A0A195FFZ1 A0A195DUW5 E2AIN0 A0A151XIL3 A0A026WXN1 A0A0K8W5Q0 K7J0F9 A0A1I8NCT4 A0A034WNE9 F4WN38 A0A2A3E2L8 A0A0M3QXG1 B4M5D3 B4JUP5 E2C4Z5 A0A0A1WHJ4 W8BV78 B4KDZ5 A0A1I8PJB7
A0A2A4JM26 A0A212F480 A0A0N1IP48 A0A194Q4T7 A0A1B0FNB0 A0A1B0AJ69 A0A182YHY9 A0A1A9UWE0 A0A1A9WJ22 A0A0L0C3D7 A0A1I8MR61 A0A1I8QBZ1 W8BEW6 A0A1Y9J0M5 A0A1S4GBG5 A0A336KD98 Q7PX67 A0A182QLK4 A0A1Y9IVC5 A0A2P8YWI1 A0A1J1IMB4 A0A1Y9IS36 A0A182IYA3 Q179I1 A0A2J7PVA2 A0A182H4P8 A0A084W049 A0A182GLY2 A0A182YQ25 A0A182T454 A0A182J1H6 W5J598 A0A182FS04 A0A3Q0JLL4 A0A182TGW3 B0WFL2 A0A1B6MCG7 D6WKJ3 Q7PUC4 A0A182RA57 A0A182NQP9 A0A182KAT0 Q294Z8 A0A182PRS9 B4KDZ6 A0A3B0KP62 B4NAW4 Q5U198 B4JUP6 Q59DX6 B4M5D4 B3P4J7 A0A1W4WDJ3 A0A0M4EP47 B4PM67 B4QU29 A0A182HKE1 A0A182X9F5 A0A1J1IH72 B4HI17 A0A1S4GBB1 A0A182VFY5 A0A1Y9IVK1 A0A224XKD3 T1I4D7 A0A182MCL2 A0A182QP59 A0A2S2R2Z7 J9L9Q8 A0A2H8U2T4 A0A088ARL1 B3M0X9 A0A084W055 B4GME3 A0A0C9PJB4 A0A195FFZ1 A0A195DUW5 E2AIN0 A0A151XIL3 A0A026WXN1 A0A0K8W5Q0 K7J0F9 A0A1I8NCT4 A0A034WNE9 F4WN38 A0A2A3E2L8 A0A0M3QXG1 B4M5D3 B4JUP5 E2C4Z5 A0A0A1WHJ4 W8BV78 B4KDZ5 A0A1I8PJB7
PDB
6EG0
E-value=2.30095e-40,
Score=409
Ontologies
GO
PANTHER
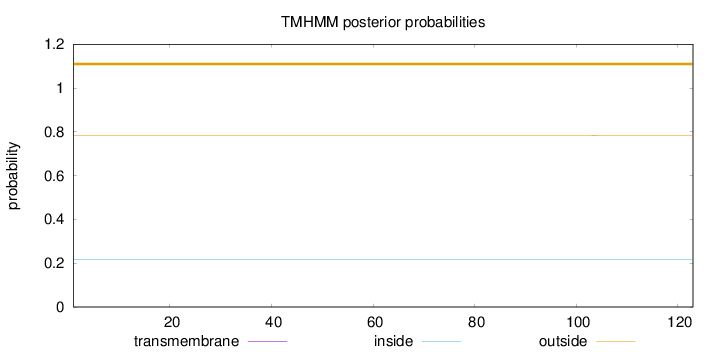
Topology
Length:
123
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00319
Exp number, first 60 AAs:
0.00135
Total prob of N-in:
0.21740
outside
1 - 123
Population Genetic Test Statistics
Pi
208.634561
Theta
184.342095
Tajima's D
0.246984
CLR
0.006526
CSRT
0.445077746112694
Interpretation
Uncertain
