Gene
KWMTBOMO12538
Annotation
PREDICTED:_uncharacterized_protein_LOC106115416_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 4.591
Sequence
CDS
ATGGATCAAGACCGTAAGAATAACTGGAGTTGTCAGGAATGTCGTAGTAAGATACCGAAATCTGACAACACAAATACTCCTGCTAGAGCAGTGAGTCGATCCGCCACTTCTGATGAAGATAACGGTCTTGGAGAAGAAAGAAATATTACTTTGCGAACAAAAAAACAGAGCTCGAAATGTGACAGCGATGGATCCTACGTTACTGAAGGACATTTACGTATCATTATTAAACAGGAGATAACAAAAACTATCAAACAACTAGTTTCTGAACACTTATCGAATATTTCCCAACAAATAGCTGACTTCCACGAATCCTTAACATTTTTTAATAAACAATACGAGGAACTCAAGCAAATGGTAAGCGAGAGGGACGATACAATTTCTGATTTAGTTTCTAAAAACAATAATTTAACAATACAAGTGAAGAATTTGACAAATAGACTGGGGCAAGTAGAACAAAGCATGCGTTCCCCAAATATTGAGATCAATGGTATCCCTGAAAATAAATCAGAAAATTTAATAACGACTATTGAGCAGCTGGGTAGAATTGTCGGAAATCCTTTTGGCGACAGCGACATTCTCCACGTAACACGTATTGCCAAACTTAATAAGGAGAATGACCGTCCTCGCTCCGTGATTATTAAACTACGTAGTCAGCGCCGTCGCGATGAGTTGCTGGCTGCAGTTACTAAGTTCAATAAGAAGAATTTAGACAACAAATTGAATTCAGAGCATCTAGGAATCGGGGGACGTCGTGTGCCAGTATTTGTATCTGAACATCTAACGCCGATAAATAAACACTTACATGCGGCCGCTCGTAAGAAGACAAAAGATACTGGTTTTAAGTTTATATGGGTGAGGGATGGTAAAGTTTTCGTAAGGAAAAATGAGCAGAGTCACGTTATATGTATTAAAGACGAAGAAAGCTTAAAATTAATTGCCTAG
Protein
MDQDRKNNWSCQECRSKIPKSDNTNTPARAVSRSATSDEDNGLGEERNITLRTKKQSSKCDSDGSYVTEGHLRIIIKQEITKTIKQLVSEHLSNISQQIADFHESLTFFNKQYEELKQMVSERDDTISDLVSKNNNLTIQVKNLTNRLGQVEQSMRSPNIEINGIPENKSENLITTIEQLGRIVGNPFGDSDILHVTRIAKLNKENDRPRSVIIKLRSQRRRDELLAAVTKFNKKNLDNKLNSEHLGIGGRRVPVFVSEHLTPINKHLHAAARKKTKDTGFKFIWVRDGKVFVRKNEQSHVICIKDEESLKLIA
Summary
Similarity
Belongs to the cytochrome P450 family.
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Uniprot
A0A194QEB0
A0A2A4IUU8
A0A2H1VRS8
A0A2A4JC78
A0A2H1WYV3
A0A2A4IYB9
+ More
A0A2H1VP34 A0A0L7KN20 A0A0L7L1J8 A0A0L7LEU2 A0A437AS13 A0A0L7K4K4 A0A0L7LEV4 A0A0L7KQ92 A0A2H1WBH0 A0A2A4IUX2 A0A2W1BYU6 A0A2A4K1S6 A0A2A4IU35 A0A0L7K4I6 A0A0L7K3E6 A0A2H1W956 A0A2H1W117 A0A2A4JD60 A0A2H1V294 A0A0L7L0X6 A0A2A4IW47 A0A2H1VD37 A0A2H1VAR2 A0A2H1W5G4 A0A2A4J0C3 A0A2A4JBL7 A0A2W1BT87 A0A0L7K1T2 A0A0L7L635 A0A0L7KUP7 A0A0L7LTI6 A0A0L7K2U8 A0A0L7L3N9 A0A2A4IZ89 A0A2H1VJZ0 A0A212EWC1 A0A0L7L3E9 A0A437ASZ4 A0A2A4JWA7 A0A2H1X1G7 A0A194QTI2 A0A2H1VKR1 G3E4N3 A0A0L7LQ84 A0A1E1WIH3 A0A2A4IYM6 A0A0L7L8J7 A0A212F6C5 A0A1Y1JXQ1 A0A2A4JD31 A0A2A4JR78 A0A0L7LCM2 A0A2A4JUE7 A0A0L7KR27 A0A0A9WQ01 A0A0C9S1R0 A0A0L7LLD8 A0A1S3DD01 A0A2H1V922 A0A2H1X1K9 A0A2H1WNV1 A0A2H1WI81 A0A0L7KQ78 A0A2A4J0K9 W4Z7K5 A0A1S3DGR2 A0A0L7K2E3 A0A0L7LBF0 A0A0L7LG83 A0A194PTX1
A0A2H1VP34 A0A0L7KN20 A0A0L7L1J8 A0A0L7LEU2 A0A437AS13 A0A0L7K4K4 A0A0L7LEV4 A0A0L7KQ92 A0A2H1WBH0 A0A2A4IUX2 A0A2W1BYU6 A0A2A4K1S6 A0A2A4IU35 A0A0L7K4I6 A0A0L7K3E6 A0A2H1W956 A0A2H1W117 A0A2A4JD60 A0A2H1V294 A0A0L7L0X6 A0A2A4IW47 A0A2H1VD37 A0A2H1VAR2 A0A2H1W5G4 A0A2A4J0C3 A0A2A4JBL7 A0A2W1BT87 A0A0L7K1T2 A0A0L7L635 A0A0L7KUP7 A0A0L7LTI6 A0A0L7K2U8 A0A0L7L3N9 A0A2A4IZ89 A0A2H1VJZ0 A0A212EWC1 A0A0L7L3E9 A0A437ASZ4 A0A2A4JWA7 A0A2H1X1G7 A0A194QTI2 A0A2H1VKR1 G3E4N3 A0A0L7LQ84 A0A1E1WIH3 A0A2A4IYM6 A0A0L7L8J7 A0A212F6C5 A0A1Y1JXQ1 A0A2A4JD31 A0A2A4JR78 A0A0L7LCM2 A0A2A4JUE7 A0A0L7KR27 A0A0A9WQ01 A0A0C9S1R0 A0A0L7LLD8 A0A1S3DD01 A0A2H1V922 A0A2H1X1K9 A0A2H1WNV1 A0A2H1WI81 A0A0L7KQ78 A0A2A4J0K9 W4Z7K5 A0A1S3DGR2 A0A0L7K2E3 A0A0L7LBF0 A0A0L7LG83 A0A194PTX1
EMBL
KQ459054
KPJ03878.1
NWSH01007173
PCG63054.1
ODYU01004037
SOQ43507.1
+ More
NWSH01002032 PCG69366.1 ODYU01012049 SOQ58146.1 NWSH01005479 PCG64170.1 ODYU01003592 SOQ42557.1 JTDY01008545 KOB64520.1 JTDY01003535 KOB69383.1 JTDY01001361 KOB74078.1 RSAL01003019 RVE40362.1 JTDY01010989 KOB57787.1 JTDY01001411 KOB73935.1 JTDY01007401 KOB65231.1 ODYU01007238 SOQ49844.1 NWSH01006948 PCG63178.1 KZ149914 PZC78000.1 NWSH01000267 PCG77878.1 NWSH01007563 PCG62908.1 KOB57786.1 JTDY01014577 KOB52017.1 ODYU01007128 SOQ49629.1 ODYU01005672 SOQ46768.1 NWSH01001819 PCG70025.1 ODYU01000349 SOQ34960.1 JTDY01003733 KOB69075.1 NWSH01005702 PCG64035.1 ODYU01001881 SOQ38706.1 ODYU01001557 SOQ37940.1 ODYU01006462 SOQ48321.1 NWSH01004587 PCG64890.1 NWSH01002191 PCG68944.1 KZ150018 PZC74943.1 JTDY01018098 KOB51825.1 JTDY01015497 JTDY01002687 KOB51945.1 KOB70917.1 JTDY01005451 KOB66988.1 JTDY01000136 KOB78669.1 JTDY01013334 JTDY01001687 KOB52155.1 KOB73157.1 JTDY01003125 KOB70108.1 NWSH01004298 PCG65247.1 ODYU01002971 SOQ41143.1 AGBW02012032 OWR45776.1 JTDY01003264 KOB69846.1 RSAL01000599 RVE41326.1 NWSH01000452 NWSH01000043 PCG76317.1 PCG80309.1 ODYU01012716 SOQ59161.1 KQ461154 KPJ08619.1 ODYU01002850 SOQ40844.1 HQ585015 AEM44816.1 JTDY01000399 KOB77361.1 GDQN01004260 JAT86794.1 NWSH01004492 PCG65047.1 JTDY01002226 KOB71853.1 AGBW02010051 OWR49283.1 GEZM01097808 JAV54094.1 NWSH01002051 PCG69303.1 NWSH01000830 PCG73963.1 JTDY01001653 KOB73242.1 NWSH01000638 PCG75103.1 JTDY01007036 KOB65506.1 GBHO01033077 JAG10527.1 GBZX01001916 JAG90824.1 JTDY01000678 KOB76257.1 ODYU01001291 SOQ37301.1 ODYU01012701 SOQ59142.1 ODYU01009977 SOQ54751.1 ODYU01008832 SOQ52780.1 JTDY01007074 KOB65473.1 NWSH01003975 PCG65605.1 AAGJ04046517 JTDY01015045 KOB51976.1 JTDY01001811 KOB72813.1 JTDY01001253 KOB74394.1 KQ459592 KPI96861.1
NWSH01002032 PCG69366.1 ODYU01012049 SOQ58146.1 NWSH01005479 PCG64170.1 ODYU01003592 SOQ42557.1 JTDY01008545 KOB64520.1 JTDY01003535 KOB69383.1 JTDY01001361 KOB74078.1 RSAL01003019 RVE40362.1 JTDY01010989 KOB57787.1 JTDY01001411 KOB73935.1 JTDY01007401 KOB65231.1 ODYU01007238 SOQ49844.1 NWSH01006948 PCG63178.1 KZ149914 PZC78000.1 NWSH01000267 PCG77878.1 NWSH01007563 PCG62908.1 KOB57786.1 JTDY01014577 KOB52017.1 ODYU01007128 SOQ49629.1 ODYU01005672 SOQ46768.1 NWSH01001819 PCG70025.1 ODYU01000349 SOQ34960.1 JTDY01003733 KOB69075.1 NWSH01005702 PCG64035.1 ODYU01001881 SOQ38706.1 ODYU01001557 SOQ37940.1 ODYU01006462 SOQ48321.1 NWSH01004587 PCG64890.1 NWSH01002191 PCG68944.1 KZ150018 PZC74943.1 JTDY01018098 KOB51825.1 JTDY01015497 JTDY01002687 KOB51945.1 KOB70917.1 JTDY01005451 KOB66988.1 JTDY01000136 KOB78669.1 JTDY01013334 JTDY01001687 KOB52155.1 KOB73157.1 JTDY01003125 KOB70108.1 NWSH01004298 PCG65247.1 ODYU01002971 SOQ41143.1 AGBW02012032 OWR45776.1 JTDY01003264 KOB69846.1 RSAL01000599 RVE41326.1 NWSH01000452 NWSH01000043 PCG76317.1 PCG80309.1 ODYU01012716 SOQ59161.1 KQ461154 KPJ08619.1 ODYU01002850 SOQ40844.1 HQ585015 AEM44816.1 JTDY01000399 KOB77361.1 GDQN01004260 JAT86794.1 NWSH01004492 PCG65047.1 JTDY01002226 KOB71853.1 AGBW02010051 OWR49283.1 GEZM01097808 JAV54094.1 NWSH01002051 PCG69303.1 NWSH01000830 PCG73963.1 JTDY01001653 KOB73242.1 NWSH01000638 PCG75103.1 JTDY01007036 KOB65506.1 GBHO01033077 JAG10527.1 GBZX01001916 JAG90824.1 JTDY01000678 KOB76257.1 ODYU01001291 SOQ37301.1 ODYU01012701 SOQ59142.1 ODYU01009977 SOQ54751.1 ODYU01008832 SOQ52780.1 JTDY01007074 KOB65473.1 NWSH01003975 PCG65605.1 AAGJ04046517 JTDY01015045 KOB51976.1 JTDY01001811 KOB72813.1 JTDY01001253 KOB74394.1 KQ459592 KPI96861.1
Proteomes
Pfam
Interpro
IPR011011
Znf_FYVE_PHD
+ More
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR004244 Transposase_22
IPR004941 Baculo_FP
IPR036691 Endo/exonu/phosph_ase_sf
IPR002401 Cyt_P450_E_grp-I
IPR036396 Cyt_P450_sf
IPR001128 Cyt_P450
IPR017972 Cyt_P450_CS
IPR019786 Zinc_finger_PHD-type_CS
IPR001965 Znf_PHD
IPR005135 Endo/exonuclease/phosphatase
IPR002067 Mit_carrier
IPR018108 Mitochondrial_sb/sol_carrier
IPR023395 Mt_carrier_dom_sf
IPR003593 AAA+_ATPase
IPR026082 ABCA
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR001841 Znf_RING
IPR038441 THAP_Znf_sf
IPR006612 THAP_Znf
IPR021896 Transposase_37
IPR002219 PE/DAG-bd
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR004244 Transposase_22
IPR004941 Baculo_FP
IPR036691 Endo/exonu/phosph_ase_sf
IPR002401 Cyt_P450_E_grp-I
IPR036396 Cyt_P450_sf
IPR001128 Cyt_P450
IPR017972 Cyt_P450_CS
IPR019786 Zinc_finger_PHD-type_CS
IPR001965 Znf_PHD
IPR005135 Endo/exonuclease/phosphatase
IPR002067 Mit_carrier
IPR018108 Mitochondrial_sb/sol_carrier
IPR023395 Mt_carrier_dom_sf
IPR003593 AAA+_ATPase
IPR026082 ABCA
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR001841 Znf_RING
IPR038441 THAP_Znf_sf
IPR006612 THAP_Znf
IPR021896 Transposase_37
IPR002219 PE/DAG-bd
SUPFAM
ProteinModelPortal
A0A194QEB0
A0A2A4IUU8
A0A2H1VRS8
A0A2A4JC78
A0A2H1WYV3
A0A2A4IYB9
+ More
A0A2H1VP34 A0A0L7KN20 A0A0L7L1J8 A0A0L7LEU2 A0A437AS13 A0A0L7K4K4 A0A0L7LEV4 A0A0L7KQ92 A0A2H1WBH0 A0A2A4IUX2 A0A2W1BYU6 A0A2A4K1S6 A0A2A4IU35 A0A0L7K4I6 A0A0L7K3E6 A0A2H1W956 A0A2H1W117 A0A2A4JD60 A0A2H1V294 A0A0L7L0X6 A0A2A4IW47 A0A2H1VD37 A0A2H1VAR2 A0A2H1W5G4 A0A2A4J0C3 A0A2A4JBL7 A0A2W1BT87 A0A0L7K1T2 A0A0L7L635 A0A0L7KUP7 A0A0L7LTI6 A0A0L7K2U8 A0A0L7L3N9 A0A2A4IZ89 A0A2H1VJZ0 A0A212EWC1 A0A0L7L3E9 A0A437ASZ4 A0A2A4JWA7 A0A2H1X1G7 A0A194QTI2 A0A2H1VKR1 G3E4N3 A0A0L7LQ84 A0A1E1WIH3 A0A2A4IYM6 A0A0L7L8J7 A0A212F6C5 A0A1Y1JXQ1 A0A2A4JD31 A0A2A4JR78 A0A0L7LCM2 A0A2A4JUE7 A0A0L7KR27 A0A0A9WQ01 A0A0C9S1R0 A0A0L7LLD8 A0A1S3DD01 A0A2H1V922 A0A2H1X1K9 A0A2H1WNV1 A0A2H1WI81 A0A0L7KQ78 A0A2A4J0K9 W4Z7K5 A0A1S3DGR2 A0A0L7K2E3 A0A0L7LBF0 A0A0L7LG83 A0A194PTX1
A0A2H1VP34 A0A0L7KN20 A0A0L7L1J8 A0A0L7LEU2 A0A437AS13 A0A0L7K4K4 A0A0L7LEV4 A0A0L7KQ92 A0A2H1WBH0 A0A2A4IUX2 A0A2W1BYU6 A0A2A4K1S6 A0A2A4IU35 A0A0L7K4I6 A0A0L7K3E6 A0A2H1W956 A0A2H1W117 A0A2A4JD60 A0A2H1V294 A0A0L7L0X6 A0A2A4IW47 A0A2H1VD37 A0A2H1VAR2 A0A2H1W5G4 A0A2A4J0C3 A0A2A4JBL7 A0A2W1BT87 A0A0L7K1T2 A0A0L7L635 A0A0L7KUP7 A0A0L7LTI6 A0A0L7K2U8 A0A0L7L3N9 A0A2A4IZ89 A0A2H1VJZ0 A0A212EWC1 A0A0L7L3E9 A0A437ASZ4 A0A2A4JWA7 A0A2H1X1G7 A0A194QTI2 A0A2H1VKR1 G3E4N3 A0A0L7LQ84 A0A1E1WIH3 A0A2A4IYM6 A0A0L7L8J7 A0A212F6C5 A0A1Y1JXQ1 A0A2A4JD31 A0A2A4JR78 A0A0L7LCM2 A0A2A4JUE7 A0A0L7KR27 A0A0A9WQ01 A0A0C9S1R0 A0A0L7LLD8 A0A1S3DD01 A0A2H1V922 A0A2H1X1K9 A0A2H1WNV1 A0A2H1WI81 A0A0L7KQ78 A0A2A4J0K9 W4Z7K5 A0A1S3DGR2 A0A0L7K2E3 A0A0L7LBF0 A0A0L7LG83 A0A194PTX1
Ontologies
KEGG
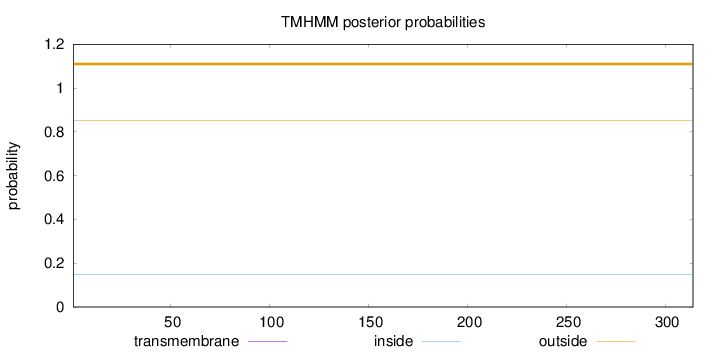
Topology
Length:
314
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.14926
outside
1 - 314
Population Genetic Test Statistics
Pi
203.747792
Theta
126.98163
Tajima's D
1.842678
CLR
92.317095
CSRT
0.857757112144393
Interpretation
Uncertain
