Gene
KWMTBOMO12536
Pre Gene Modal
BGIBMGA001456
Annotation
PREDICTED:_G_patch_domain_and_KOW_motifs-containing_protein_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 4.54
Sequence
CDS
ATGGAGGGCAAAAAAATATCTTTTGGTTTCATGAAAACGAAAAAAGCAGAAAAACCAGTCGTAGACGAAAAAAAGGACTATATAGAATGTGTAGAGGAGAAATCTATCAAAGTAGTCGGTGCGGAAGAAATCAAAGATGACACACCTTTAATAATTCCAATGAAACCTAACACGCTCATCACGACTGAGAGATTGAAGCAGATAGCCCAGAAAGTAGAGAGTGCTTTAGATGAACCGGAAGTTGTGAAGTCAGATAGTCCAAAGACTGTTGAAATTCCAGAAAATGAGACTTTAGAGCAGATGGCTGTCAGAGAATTAATGGAAGACACGAAGAAGAAGGTGAAAATTGAAACGTCAGCCATAACAGTACCGTTACCTGCTAAACCAGTTACTGATGGAGAAAAAGAGTCAACATTGGATGACTATGACTCTGTACCAATACAAGACTTTGGTATGGCAATGCTTCGTGGAATGGGATGGACACCAAACAAAGACCTGCCCAAATACAAGCAACCTCAGTTAAGGCCTAAAGGCCTAGGACTTGGTGCCGACAAAGTTGTCTCAGATAAAAAGAAAACATCTAAAGACAAAAATGGTGAAGAATTAACAATCGTGAAGAACTCATTTGTGAAAATTACAACTGGGAAATACAGTGGATATTATGGAAAGGTGCTTAGCCTCGATGAAGACAACGGCAGAGCTATGGTCAATATAGTGATGAAAAATGAAACTGTCAATCTAAGCGAGTTCATGATGCATCCGGTCACAAAGTCCGAATTCGACAAGGAATCCAAAGTTATAAATGCAGACAAGTACGAAAAATACAAGAAAGATGAAAAAGTTCCTCAATCGTCAAAAGAAAAAGATATAGAGAGTTCACCTAAAAGAGAAACGAAGAAAGAAAAGTATGAACATACCGACAGGCGTGAAAGAGAAGATTGGTCAAATGGTAAAAATAGAAAAGAGGATAGACATAGGAGAGACTCGTCAAGCACCGGTGAAGAGAGATCTCAGAAGAAAAAGAAGCACAAAAAAAGATACAGCAGCAACGAATCTTTAAGTTCTCAATCAGAGGATGAGAGGAGGCGGAGGAAGAAAGACAAGAAAAAGCGTGAAAGAAGCGTGGATTCTGAAGATGAATATAAAAAGAATAAAGGTAGCGCTACACCGGACGGCAAACGAGATATCGATAAGAAGAGGAAGGGAAAAAAGAAGAAAAGAGACAGGGATCGTTCACCGAATTATAAAAAACATAGAAAATAA
Protein
MEGKKISFGFMKTKKAEKPVVDEKKDYIECVEEKSIKVVGAEEIKDDTPLIIPMKPNTLITTERLKQIAQKVESALDEPEVVKSDSPKTVEIPENETLEQMAVRELMEDTKKKVKIETSAITVPLPAKPVTDGEKESTLDDYDSVPIQDFGMAMLRGMGWTPNKDLPKYKQPQLRPKGLGLGADKVVSDKKKTSKDKNGEELTIVKNSFVKITTGKYSGYYGKVLSLDEDNGRAMVNIVMKNETVNLSEFMMHPVTKSEFDKESKVINADKYEKYKKDEKVPQSSKEKDIESSPKRETKKEKYEHTDRREREDWSNGKNRKEDRHRRDSSSTGEERSQKKKKHKKRYSSNESLSSQSEDERRRRKKDKKKRERSVDSEDEYKKNKGSATPDGKRDIDKKRKGKKKKRDRDRSPNYKKHRK
Summary
Uniprot
H9IW25
A0A437BLX6
A0A2A4IVX3
A0A2H1W5J5
A0A212FH18
A0A0N0PB27
+ More
A0A194Q8C2 A0A0M9A5N5 A0A0L7R696 A0A310S9I2 A0A088A403 A0A3L8DAI1 A0A154NVZ0 A0A1Y1N458 A0A2A3EM79 A0A026WWQ1 E2C501 K7J461 A0A1Q3G186 A0A1J1I1F6 A0A1Q3G114 A0A195FQ43 A0A195D9R8 A0A158NUY3 A0A0V0G878 A0A151WSZ1 A0A2M3Z8T1 A0A1I8JST4 N6T3G2 A0A2M4BP20 A0A2M4BQG0 A0A195CPW0 A0A2M4BRA2 A0A2M4CIB7 A0A2M4CJL8 A0A2M3Z908 F4WXK4 A0A1B6MA96 A0A0A9XGZ5 A0A0K8T946 A0A151IH82 B4JG87 A0A146LG24 A0A2M4BNC8 A0A1W4U755 A0A195BVI0 B4NJH7 W8BI72 B4G4A4 Q8SZD8 Q297K1 Q9VES0 B4IBS0 D1MDC0 A0A1B6HES7 B3M2K7 B4KAH8
A0A194Q8C2 A0A0M9A5N5 A0A0L7R696 A0A310S9I2 A0A088A403 A0A3L8DAI1 A0A154NVZ0 A0A1Y1N458 A0A2A3EM79 A0A026WWQ1 E2C501 K7J461 A0A1Q3G186 A0A1J1I1F6 A0A1Q3G114 A0A195FQ43 A0A195D9R8 A0A158NUY3 A0A0V0G878 A0A151WSZ1 A0A2M3Z8T1 A0A1I8JST4 N6T3G2 A0A2M4BP20 A0A2M4BQG0 A0A195CPW0 A0A2M4BRA2 A0A2M4CIB7 A0A2M4CJL8 A0A2M3Z908 F4WXK4 A0A1B6MA96 A0A0A9XGZ5 A0A0K8T946 A0A151IH82 B4JG87 A0A146LG24 A0A2M4BNC8 A0A1W4U755 A0A195BVI0 B4NJH7 W8BI72 B4G4A4 Q8SZD8 Q297K1 Q9VES0 B4IBS0 D1MDC0 A0A1B6HES7 B3M2K7 B4KAH8
Pubmed
EMBL
BABH01006073
BABH01006074
BABH01006075
RSAL01000035
RVE51315.1
NWSH01005791
+ More
PCG63945.1 ODYU01006483 SOQ48359.1 AGBW02008561 OWR53032.1 KQ461068 KPJ09898.1 KQ459564 KPI99655.1 KQ435728 KOX77740.1 KQ414648 KOC66301.1 KQ762962 OAD55173.1 QOIP01000010 RLU17346.1 KQ434772 KZC03855.1 GEZM01013319 GEZM01013318 JAV92663.1 KZ288219 PBC32306.1 KK107079 EZA60121.1 GL452712 EFN76982.1 AAZX01013336 GFDL01001493 JAV33552.1 CVRI01000038 CRK94160.1 GFDL01001558 JAV33487.1 KQ981335 KYN42598.1 KQ981082 KYN09616.1 ADTU01026824 GECL01001777 JAP04347.1 KQ982769 KYQ50911.1 GGFM01004198 MBW24949.1 APGK01053973 KB741240 KB632257 ENN72098.1 ERL90787.1 GGFJ01005387 MBW54528.1 GGFJ01006158 MBW55299.1 KQ977444 KYN02746.1 GGFJ01006157 MBW55298.1 GGFL01000909 MBW65087.1 GGFL01000910 MBW65088.1 GGFM01004266 MBW25017.1 GL888425 EGI61074.1 GEBQ01007125 GEBQ01004519 JAT32852.1 JAT35458.1 GBHO01024355 JAG19249.1 GBRD01004136 JAG61685.1 KQ977637 KYN01127.1 CH916369 EDV93654.1 GDHC01012707 JAQ05922.1 GGFJ01005439 MBW54580.1 KQ976401 KYM92629.1 CH964272 EDW83901.1 GAMC01013615 GAMC01013614 JAB92941.1 CH479179 EDW25142.1 AY070940 AAL48562.1 CM000070 EAL28204.1 AE014297 BT150346 AAF55349.1 AGW30433.1 CH480827 EDW44828.1 GU132453 ACZ50718.1 GECU01034516 JAS73190.1 CH902617 EDV42328.1 CH933806 EDW15691.1
PCG63945.1 ODYU01006483 SOQ48359.1 AGBW02008561 OWR53032.1 KQ461068 KPJ09898.1 KQ459564 KPI99655.1 KQ435728 KOX77740.1 KQ414648 KOC66301.1 KQ762962 OAD55173.1 QOIP01000010 RLU17346.1 KQ434772 KZC03855.1 GEZM01013319 GEZM01013318 JAV92663.1 KZ288219 PBC32306.1 KK107079 EZA60121.1 GL452712 EFN76982.1 AAZX01013336 GFDL01001493 JAV33552.1 CVRI01000038 CRK94160.1 GFDL01001558 JAV33487.1 KQ981335 KYN42598.1 KQ981082 KYN09616.1 ADTU01026824 GECL01001777 JAP04347.1 KQ982769 KYQ50911.1 GGFM01004198 MBW24949.1 APGK01053973 KB741240 KB632257 ENN72098.1 ERL90787.1 GGFJ01005387 MBW54528.1 GGFJ01006158 MBW55299.1 KQ977444 KYN02746.1 GGFJ01006157 MBW55298.1 GGFL01000909 MBW65087.1 GGFL01000910 MBW65088.1 GGFM01004266 MBW25017.1 GL888425 EGI61074.1 GEBQ01007125 GEBQ01004519 JAT32852.1 JAT35458.1 GBHO01024355 JAG19249.1 GBRD01004136 JAG61685.1 KQ977637 KYN01127.1 CH916369 EDV93654.1 GDHC01012707 JAQ05922.1 GGFJ01005439 MBW54580.1 KQ976401 KYM92629.1 CH964272 EDW83901.1 GAMC01013615 GAMC01013614 JAB92941.1 CH479179 EDW25142.1 AY070940 AAL48562.1 CM000070 EAL28204.1 AE014297 BT150346 AAF55349.1 AGW30433.1 CH480827 EDW44828.1 GU132453 ACZ50718.1 GECU01034516 JAS73190.1 CH902617 EDV42328.1 CH933806 EDW15691.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000053268
+ More
UP000053105 UP000053825 UP000005203 UP000279307 UP000076502 UP000242457 UP000053097 UP000008237 UP000002358 UP000183832 UP000078541 UP000078492 UP000005205 UP000075809 UP000069272 UP000019118 UP000030742 UP000078542 UP000007755 UP000001070 UP000192221 UP000078540 UP000007798 UP000008744 UP000001819 UP000000803 UP000001292 UP000007801 UP000009192
UP000053105 UP000053825 UP000005203 UP000279307 UP000076502 UP000242457 UP000053097 UP000008237 UP000002358 UP000183832 UP000078541 UP000078492 UP000005205 UP000075809 UP000069272 UP000019118 UP000030742 UP000078542 UP000007755 UP000001070 UP000192221 UP000078540 UP000007798 UP000008744 UP000001819 UP000000803 UP000001292 UP000007801 UP000009192
Interpro
Gene 3D
ProteinModelPortal
H9IW25
A0A437BLX6
A0A2A4IVX3
A0A2H1W5J5
A0A212FH18
A0A0N0PB27
+ More
A0A194Q8C2 A0A0M9A5N5 A0A0L7R696 A0A310S9I2 A0A088A403 A0A3L8DAI1 A0A154NVZ0 A0A1Y1N458 A0A2A3EM79 A0A026WWQ1 E2C501 K7J461 A0A1Q3G186 A0A1J1I1F6 A0A1Q3G114 A0A195FQ43 A0A195D9R8 A0A158NUY3 A0A0V0G878 A0A151WSZ1 A0A2M3Z8T1 A0A1I8JST4 N6T3G2 A0A2M4BP20 A0A2M4BQG0 A0A195CPW0 A0A2M4BRA2 A0A2M4CIB7 A0A2M4CJL8 A0A2M3Z908 F4WXK4 A0A1B6MA96 A0A0A9XGZ5 A0A0K8T946 A0A151IH82 B4JG87 A0A146LG24 A0A2M4BNC8 A0A1W4U755 A0A195BVI0 B4NJH7 W8BI72 B4G4A4 Q8SZD8 Q297K1 Q9VES0 B4IBS0 D1MDC0 A0A1B6HES7 B3M2K7 B4KAH8
A0A194Q8C2 A0A0M9A5N5 A0A0L7R696 A0A310S9I2 A0A088A403 A0A3L8DAI1 A0A154NVZ0 A0A1Y1N458 A0A2A3EM79 A0A026WWQ1 E2C501 K7J461 A0A1Q3G186 A0A1J1I1F6 A0A1Q3G114 A0A195FQ43 A0A195D9R8 A0A158NUY3 A0A0V0G878 A0A151WSZ1 A0A2M3Z8T1 A0A1I8JST4 N6T3G2 A0A2M4BP20 A0A2M4BQG0 A0A195CPW0 A0A2M4BRA2 A0A2M4CIB7 A0A2M4CJL8 A0A2M3Z908 F4WXK4 A0A1B6MA96 A0A0A9XGZ5 A0A0K8T946 A0A151IH82 B4JG87 A0A146LG24 A0A2M4BNC8 A0A1W4U755 A0A195BVI0 B4NJH7 W8BI72 B4G4A4 Q8SZD8 Q297K1 Q9VES0 B4IBS0 D1MDC0 A0A1B6HES7 B3M2K7 B4KAH8
Ontologies
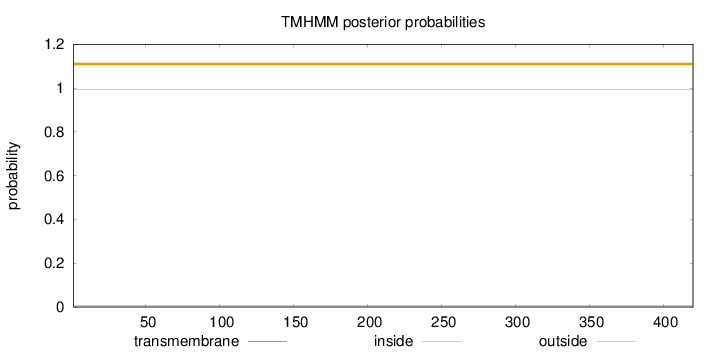
Topology
Length:
420
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00530
outside
1 - 420
Population Genetic Test Statistics
Pi
255.938628
Theta
160.946519
Tajima's D
2.005706
CLR
0.179801
CSRT
0.88325583720814
Interpretation
Uncertain
