Gene
KWMTBOMO12534
Pre Gene Modal
BGIBMGA001457
Annotation
PREDICTED:_chromatin_assembly_factor_1_subunit_A_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.246
Sequence
CDS
ATGAGATTAGCACCTACAACTAGAGCTGAATTGAGTGATGATAAGAGACATCAGTTGGACAATTTTATTAAGGAACAGAAGTGCAAAGAACATGAACTATATCTTAAATGCCTTAAGGATGGAAACAGTAAACCTCTTTCTACTGGCAAGACCTGGCCTCTAAGTGACAAAGACGATGATGTTATGATTATTGAAGATGAACTGCCACCAATAGATGCAGAAGATGAAATTGTAGCTTGCGACCAAGCACCTCGTGAAAAGCTTAGACCTAAACTATTAAGCTTCCATGAAAACCGACGGCCTCCATATTGGGGAACATGGAGAAAGAAAACCACATTTGTCAAACCTAGAAAGCCATTTGGACAGGATGAGAAACAACTTGACTACGAAGTCGATAGTGATGAGGAATGGGAAGAAGAACAGGAGGGTGAAAGCATTGATGGTAGTGCTGTTGGTAGTGATGACGAGCAAGATGGCGATGAGTATGAAGTAGATAATGAAGTGTTTGTACCGCACGGATATCTTAGTGATGAGGAAGCAACTATGGAAGATGATGACGTACTATCACTGTCCCCTGAAACACAGAAGGCGAGGCTCAAACATTTAGAGGATGAATTTGAAACTGAAATGAAGAAACCAACCGAAAAACTGAAACCACGCATGTATGGCCCACTTTGGGAAAGTAGTGATGGTGGTAAGCCTAGTAAATGCGTTGACGCTTTATGGAATTACTTCTCGAAACTAGCCATGGTTATTGATGATCCAACGCCTTTCTTACAACCTTCGAGTGAACCTGACGAACAAGAGAAAAAAAGAGTTAAGAAAAAGAAACCTGTTAATTCTGAAAACAGTGAACAAAACAATAAATCTGAAAAAAAGAAAAAGCCCAAATCAGAGGAAAAAGAAGGAAAATCGCCTAAAGTTGAAAACAAAAAGATTGGTCCCGAAAAAAAGAATCAGCCTGGCATAAATATGTTTTTAACTAAATTAAAGTCTTCATGA
Protein
MRLAPTTRAELSDDKRHQLDNFIKEQKCKEHELYLKCLKDGNSKPLSTGKTWPLSDKDDDVMIIEDELPPIDAEDEIVACDQAPREKLRPKLLSFHENRRPPYWGTWRKKTTFVKPRKPFGQDEKQLDYEVDSDEEWEEEQEGESIDGSAVGSDDEQDGDEYEVDNEVFVPHGYLSDEEATMEDDDVLSLSPETQKARLKHLEDEFETEMKKPTEKLKPRMYGPLWESSDGGKPSKCVDALWNYFSKLAMVIDDPTPFLQPSSEPDEQEKKRVKKKKPVNSENSEQNNKSEKKKKPKSEEKEGKSPKVENKKIGPEKKNQPGINMFLTKLKSS
Summary
Uniprot
H9IW26
A0A437BLF8
A0A2A4K782
A0A0N0PBB8
A0A2H1WD95
A0A194Q3S1
+ More
A0A212FH17 A0A0L7KVZ4 A0A1B0GLP8 A0A0A1XTC1 A0A034V6A1 A0A0K8W2X7 A0A0K8W630 A0A0K8TZA6 A0A0K8V218 A0A1B6E528 W8C4T6 A0A1B6EFK6 Q16L92 A0A1B0DMK4 A0A1I8Q3X6 A0A1B6MTG4 A0A0L0C4L2 A0A067R7P4 A0A1Q3EW88 A0A1Q3EWD0 A0A1B6H127 A0A1J1IY94 A0A1I8MLS0 A0A182XNW0 A0A1S4GBK2 A0A1B0BVF3 A0A1A9Y614 A0A182V8N0 J9JRC5 A0A182HK09 A0A1A9VDL1 A0A182PR41 A0A139WAU3 A0A139WAW7 A0A1W4WWR7 A0A182YQL1 A0A1W4X7F0 A0A182NR31 A0A1W4WWT7 A0A1W4X667 A0A182FB95 A0A2S2NVR4 A0A2S2QA13 A0A336L2K3 A0A182QTZ9 A0A084W099 A0A336MLL4 A0A182VWB2 N6TSU2 U5EQB6 U5EW18 W5JTJ5 A0A195F9I8 A0A146LFT6 A0A0A9ZFD3 U4UK60 A0A2M4CH85 A0A182R534 A0A182MFR2 A0A158NFA2 A0A232EIC1 K7J466 Q7PUJ4 A0A1A9WVU9 A0A151WHZ7 A0A1Y1KEG2 A0A1Y1KGX8 A0A1Y1KF04 T1JHB3 A0A023EXX9 A0A026W042 A0A3L8DFG5 A0A069DXM0 E9H2T6 A0A0M4EWV9 A0A0P6FUL8 A0A0P5JBR8 A0A162DEU8 A0A0P4Z612 A0A0P5BCX4 A0A0P6GK51 A0A0N8AUP6 A0A0P5X2T2
A0A212FH17 A0A0L7KVZ4 A0A1B0GLP8 A0A0A1XTC1 A0A034V6A1 A0A0K8W2X7 A0A0K8W630 A0A0K8TZA6 A0A0K8V218 A0A1B6E528 W8C4T6 A0A1B6EFK6 Q16L92 A0A1B0DMK4 A0A1I8Q3X6 A0A1B6MTG4 A0A0L0C4L2 A0A067R7P4 A0A1Q3EW88 A0A1Q3EWD0 A0A1B6H127 A0A1J1IY94 A0A1I8MLS0 A0A182XNW0 A0A1S4GBK2 A0A1B0BVF3 A0A1A9Y614 A0A182V8N0 J9JRC5 A0A182HK09 A0A1A9VDL1 A0A182PR41 A0A139WAU3 A0A139WAW7 A0A1W4WWR7 A0A182YQL1 A0A1W4X7F0 A0A182NR31 A0A1W4WWT7 A0A1W4X667 A0A182FB95 A0A2S2NVR4 A0A2S2QA13 A0A336L2K3 A0A182QTZ9 A0A084W099 A0A336MLL4 A0A182VWB2 N6TSU2 U5EQB6 U5EW18 W5JTJ5 A0A195F9I8 A0A146LFT6 A0A0A9ZFD3 U4UK60 A0A2M4CH85 A0A182R534 A0A182MFR2 A0A158NFA2 A0A232EIC1 K7J466 Q7PUJ4 A0A1A9WVU9 A0A151WHZ7 A0A1Y1KEG2 A0A1Y1KGX8 A0A1Y1KF04 T1JHB3 A0A023EXX9 A0A026W042 A0A3L8DFG5 A0A069DXM0 E9H2T6 A0A0M4EWV9 A0A0P6FUL8 A0A0P5JBR8 A0A162DEU8 A0A0P4Z612 A0A0P5BCX4 A0A0P6GK51 A0A0N8AUP6 A0A0P5X2T2
Pubmed
EMBL
BABH01006068
BABH01006069
BABH01006070
RSAL01000035
RVE51312.1
NWSH01000083
+ More
PCG79788.1 KQ461068 KPJ09896.1 ODYU01007878 SOQ51033.1 KQ459564 KPI99654.1 AGBW02008561 OWR53028.1 JTDY01005167 KOB67300.1 AJWK01009757 GBXI01000067 JAD14225.1 GAKP01020126 JAC38826.1 GDHF01006853 JAI45461.1 GDHF01005805 JAI46509.1 GDHF01032899 JAI19415.1 GDHF01019350 JAI32964.1 GEDC01004291 JAS33007.1 GAMC01005289 JAC01267.1 GEDC01000581 JAS36717.1 CH477914 EAT35088.1 AJVK01016861 GEBQ01000757 JAT39220.1 JRES01000911 KNC27308.1 KK852643 KDR19550.1 GFDL01015474 JAV19571.1 GFDL01015469 JAV19576.1 GECZ01001389 JAS68380.1 CVRI01000064 CRL05064.1 AAAB01008987 JXJN01021262 ABLF02018420 APCN01000811 KQ971379 KYB25056.1 KYB25055.1 GGMR01008692 MBY21311.1 GGMS01005362 MBY74565.1 UFQS01000878 UFQT01000878 SSX07440.1 SSX27780.1 AXCN02001479 ATLV01019105 KE525262 KFB43643.1 UFQS01001415 UFQT01001415 SSX10769.1 SSX30451.1 APGK01055861 KB741270 ENN71411.1 GANO01003341 JAB56530.1 GANO01000748 JAB59123.1 ADMH02000120 ETN67692.1 KQ981720 KYN37270.1 GDHC01012200 JAQ06429.1 GBHO01000923 GBHO01000922 JAG42681.1 JAG42682.1 KB632390 ERL94464.1 GGFL01000485 MBW64663.1 AXCM01001186 AXCM01001187 ADTU01013851 ADTU01013852 NNAY01004295 OXU18093.1 AAZX01011384 EAA01417.6 KQ983106 KYQ47440.1 GEZM01088717 GEZM01088713 JAV57995.1 GEZM01088715 JAV57997.1 GEZM01088716 JAV57996.1 JH431527 GBBI01004547 JAC14165.1 KK107519 EZA49423.1 QOIP01000009 RLU19046.1 GBGD01000442 JAC88447.1 GL732587 EFX73839.1 CP012528 ALC50022.1 GDIQ01042503 JAN52234.1 GDIQ01227326 JAK24399.1 LRGB01001663 KZS10844.1 GDIP01221782 JAJ01620.1 GDIP01186275 JAJ37127.1 GDIQ01032265 JAN62472.1 GDIQ01241091 JAK10634.1 GDIP01078533 JAM25182.1
PCG79788.1 KQ461068 KPJ09896.1 ODYU01007878 SOQ51033.1 KQ459564 KPI99654.1 AGBW02008561 OWR53028.1 JTDY01005167 KOB67300.1 AJWK01009757 GBXI01000067 JAD14225.1 GAKP01020126 JAC38826.1 GDHF01006853 JAI45461.1 GDHF01005805 JAI46509.1 GDHF01032899 JAI19415.1 GDHF01019350 JAI32964.1 GEDC01004291 JAS33007.1 GAMC01005289 JAC01267.1 GEDC01000581 JAS36717.1 CH477914 EAT35088.1 AJVK01016861 GEBQ01000757 JAT39220.1 JRES01000911 KNC27308.1 KK852643 KDR19550.1 GFDL01015474 JAV19571.1 GFDL01015469 JAV19576.1 GECZ01001389 JAS68380.1 CVRI01000064 CRL05064.1 AAAB01008987 JXJN01021262 ABLF02018420 APCN01000811 KQ971379 KYB25056.1 KYB25055.1 GGMR01008692 MBY21311.1 GGMS01005362 MBY74565.1 UFQS01000878 UFQT01000878 SSX07440.1 SSX27780.1 AXCN02001479 ATLV01019105 KE525262 KFB43643.1 UFQS01001415 UFQT01001415 SSX10769.1 SSX30451.1 APGK01055861 KB741270 ENN71411.1 GANO01003341 JAB56530.1 GANO01000748 JAB59123.1 ADMH02000120 ETN67692.1 KQ981720 KYN37270.1 GDHC01012200 JAQ06429.1 GBHO01000923 GBHO01000922 JAG42681.1 JAG42682.1 KB632390 ERL94464.1 GGFL01000485 MBW64663.1 AXCM01001186 AXCM01001187 ADTU01013851 ADTU01013852 NNAY01004295 OXU18093.1 AAZX01011384 EAA01417.6 KQ983106 KYQ47440.1 GEZM01088717 GEZM01088713 JAV57995.1 GEZM01088715 JAV57997.1 GEZM01088716 JAV57996.1 JH431527 GBBI01004547 JAC14165.1 KK107519 EZA49423.1 QOIP01000009 RLU19046.1 GBGD01000442 JAC88447.1 GL732587 EFX73839.1 CP012528 ALC50022.1 GDIQ01042503 JAN52234.1 GDIQ01227326 JAK24399.1 LRGB01001663 KZS10844.1 GDIP01221782 JAJ01620.1 GDIP01186275 JAJ37127.1 GDIQ01032265 JAN62472.1 GDIQ01241091 JAK10634.1 GDIP01078533 JAM25182.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000092461 UP000008820 UP000092462 UP000095300 UP000037069 UP000027135 UP000183832 UP000095301 UP000076407 UP000092460 UP000092443 UP000075903 UP000007819 UP000075840 UP000078200 UP000075885 UP000007266 UP000192223 UP000076408 UP000075884 UP000069272 UP000075886 UP000030765 UP000075920 UP000019118 UP000000673 UP000078541 UP000030742 UP000075900 UP000075883 UP000005205 UP000215335 UP000002358 UP000007062 UP000091820 UP000075809 UP000053097 UP000279307 UP000000305 UP000092553 UP000076858
UP000037510 UP000092461 UP000008820 UP000092462 UP000095300 UP000037069 UP000027135 UP000183832 UP000095301 UP000076407 UP000092460 UP000092443 UP000075903 UP000007819 UP000075840 UP000078200 UP000075885 UP000007266 UP000192223 UP000076408 UP000075884 UP000069272 UP000075886 UP000030765 UP000075920 UP000019118 UP000000673 UP000078541 UP000030742 UP000075900 UP000075883 UP000005205 UP000215335 UP000002358 UP000007062 UP000091820 UP000075809 UP000053097 UP000279307 UP000000305 UP000092553 UP000076858
PRIDE
ProteinModelPortal
H9IW26
A0A437BLF8
A0A2A4K782
A0A0N0PBB8
A0A2H1WD95
A0A194Q3S1
+ More
A0A212FH17 A0A0L7KVZ4 A0A1B0GLP8 A0A0A1XTC1 A0A034V6A1 A0A0K8W2X7 A0A0K8W630 A0A0K8TZA6 A0A0K8V218 A0A1B6E528 W8C4T6 A0A1B6EFK6 Q16L92 A0A1B0DMK4 A0A1I8Q3X6 A0A1B6MTG4 A0A0L0C4L2 A0A067R7P4 A0A1Q3EW88 A0A1Q3EWD0 A0A1B6H127 A0A1J1IY94 A0A1I8MLS0 A0A182XNW0 A0A1S4GBK2 A0A1B0BVF3 A0A1A9Y614 A0A182V8N0 J9JRC5 A0A182HK09 A0A1A9VDL1 A0A182PR41 A0A139WAU3 A0A139WAW7 A0A1W4WWR7 A0A182YQL1 A0A1W4X7F0 A0A182NR31 A0A1W4WWT7 A0A1W4X667 A0A182FB95 A0A2S2NVR4 A0A2S2QA13 A0A336L2K3 A0A182QTZ9 A0A084W099 A0A336MLL4 A0A182VWB2 N6TSU2 U5EQB6 U5EW18 W5JTJ5 A0A195F9I8 A0A146LFT6 A0A0A9ZFD3 U4UK60 A0A2M4CH85 A0A182R534 A0A182MFR2 A0A158NFA2 A0A232EIC1 K7J466 Q7PUJ4 A0A1A9WVU9 A0A151WHZ7 A0A1Y1KEG2 A0A1Y1KGX8 A0A1Y1KF04 T1JHB3 A0A023EXX9 A0A026W042 A0A3L8DFG5 A0A069DXM0 E9H2T6 A0A0M4EWV9 A0A0P6FUL8 A0A0P5JBR8 A0A162DEU8 A0A0P4Z612 A0A0P5BCX4 A0A0P6GK51 A0A0N8AUP6 A0A0P5X2T2
A0A212FH17 A0A0L7KVZ4 A0A1B0GLP8 A0A0A1XTC1 A0A034V6A1 A0A0K8W2X7 A0A0K8W630 A0A0K8TZA6 A0A0K8V218 A0A1B6E528 W8C4T6 A0A1B6EFK6 Q16L92 A0A1B0DMK4 A0A1I8Q3X6 A0A1B6MTG4 A0A0L0C4L2 A0A067R7P4 A0A1Q3EW88 A0A1Q3EWD0 A0A1B6H127 A0A1J1IY94 A0A1I8MLS0 A0A182XNW0 A0A1S4GBK2 A0A1B0BVF3 A0A1A9Y614 A0A182V8N0 J9JRC5 A0A182HK09 A0A1A9VDL1 A0A182PR41 A0A139WAU3 A0A139WAW7 A0A1W4WWR7 A0A182YQL1 A0A1W4X7F0 A0A182NR31 A0A1W4WWT7 A0A1W4X667 A0A182FB95 A0A2S2NVR4 A0A2S2QA13 A0A336L2K3 A0A182QTZ9 A0A084W099 A0A336MLL4 A0A182VWB2 N6TSU2 U5EQB6 U5EW18 W5JTJ5 A0A195F9I8 A0A146LFT6 A0A0A9ZFD3 U4UK60 A0A2M4CH85 A0A182R534 A0A182MFR2 A0A158NFA2 A0A232EIC1 K7J466 Q7PUJ4 A0A1A9WVU9 A0A151WHZ7 A0A1Y1KEG2 A0A1Y1KGX8 A0A1Y1KF04 T1JHB3 A0A023EXX9 A0A026W042 A0A3L8DFG5 A0A069DXM0 E9H2T6 A0A0M4EWV9 A0A0P6FUL8 A0A0P5JBR8 A0A162DEU8 A0A0P4Z612 A0A0P5BCX4 A0A0P6GK51 A0A0N8AUP6 A0A0P5X2T2
Ontologies
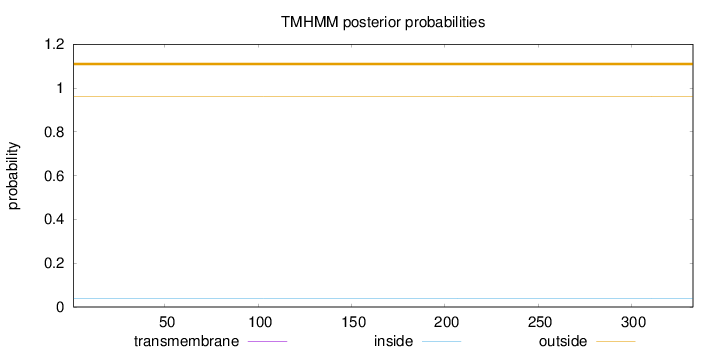
Topology
Length:
333
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03741
outside
1 - 333
Population Genetic Test Statistics
Pi
136.257168
Theta
167.002111
Tajima's D
-0.520518
CLR
0.816405
CSRT
0.23798810059497
Interpretation
Uncertain
