Gene
KWMTBOMO12532
Pre Gene Modal
BGIBMGA001513
Annotation
PREDICTED:_furin-like_convetase_isoform_X1_[Bombyx_mori]
Full name
Furin-like protease 1, isoform 1-CRR
Alternative Name
Kex2-like endoprotease 1
dKLIP-1
dKLIP-1
Location in the cell
Extracellular Reliability : 1.843 Nuclear Reliability : 1.348
Sequence
CDS
ATGTGGGGTGAAAATCCGCTGGGCGAGTGGCAACTGGAGGTCACAAATGAAGGCCGCTACATGGAAGCCGGCACTTTAACTCAATGGGAGCTGATTTTCTACGGAACAGAGACACCGGCTCAGGAGCAAGACGTGTCACCGGAGACCAACTCTTTGGGTGAAAATTCTCGGGTTGACACGAAACCTTGGACCTTGTCTGAGCCTGAATCTATTGAAGAAGTAAGACAGAACGCCATTGATGATGATCTGGCTCTCGTTTGGCATGACTCTCAAACGATCCGTGAAGAGAATCCAGTCGGTGCTGGAGACGTCGACACGGGACAGTACGCCGCTAGCGCCGCGGGCTGCGCCACCCACGCTCCGCAGCCGCCGCATACCTGCATAGAATGCGCGAAGGGCTTGCATTTGTACAACGGGCGGTGCTACTCACGTTGTCCGGACGGTACGTACGCCAATGAGATCTCGATGGAACGGAGCTCCAGAAGACGTAACCTTACAATTTTTTCTGAGGGTTCCCTTAGCAAGCGGCAAGACGGGTCACTAAAATCGTCAGCTCTGGAGGCTTTAGACATGGAACCGTATGCGAACTCAACTAAAGATCCTTTGATTTGCCTACCATGCCATTACACCTGTGCCACGTGCGCAGGTCCTCACGACAGTCAATGTGTATCTTGTCTAGACGACGCGGAGTTGTTCAACTCGACAGATTCGGTTCTCAAGTTCTATTGCTACCCAAAAAAGGTCGTGTCACAGATAAGCGACGTCAACTGGCATTATAGATTGAATGTCGTCCTTTCCCTAGTTTTGTTCTGTATTTGTTTTATTTCTCTTTATTTTATTATATCGTGGACCCTGAAATGGTTTTACGGTACGAACAATTACAATTCGAACATCGCGTACAACAAATTATCGTCGGACGAAAAACAGCAAAGCGCTTCCGAGGTCGAGGAAGAGATCCGGTTGGCTCTGAAGGATTACAGTGAGGGCGAGACAGAAGATGATCTAAATTTATAG
Protein
MWGENPLGEWQLEVTNEGRYMEAGTLTQWELIFYGTETPAQEQDVSPETNSLGENSRVDTKPWTLSEPESIEEVRQNAIDDDLALVWHDSQTIREENPVGAGDVDTGQYAASAAGCATHAPQPPHTCIECAKGLHLYNGRCYSRCPDGTYANEISMERSSRRRNLTIFSEGSLSKRQDGSLKSSALEALDMEPYANSTKDPLICLPCHYTCATCAGPHDSQCVSCLDDAELFNSTDSVLKFYCYPKKVVSQISDVNWHYRLNVVLSLVLFCICFISLYFIISWTLKWFYGTNNYNSNIAYNKLSSDEKQQSASEVEEEIRLALKDYSEGETEDDLNL
Summary
Description
Furin is likely to represent the ubiquitous endoprotease activity within constitutive secretory pathways and capable of cleavage at the RX(K/R)R consensus motif.
Catalytic Activity
Release of mature proteins from their proproteins by cleavage of -Arg-Xaa-Yaa-Arg-|-Zaa- bonds, where Xaa can be any amino acid and Yaa is Arg or Lys. Releases albumin, complement component C3 and von Willebrand factor from their respective precursors.
Cofactor
Ca(2+)
Similarity
Belongs to the peptidase S8 family.
Belongs to the peptidase S8 family. Furin subfamily.
Belongs to the peptidase S8 family. Furin subfamily.
Keywords
Alternative splicing
Cleavage on pair of basic residues
Complete proteome
Disulfide bond
Glycoprotein
Golgi apparatus
Hydrolase
Membrane
Protease
Reference proteome
Serine protease
Signal
Transmembrane
Transmembrane helix
Zymogen
Feature
chain Furin-like protease 1, isoform 1-CRR
Uniprot
A0A1E1WNT8
A0A2A4JTN4
A0A437BLV5
A0A2H1WYJ9
A0A2A4JSW4
A0A212F132
+ More
A0A194PYH4 A0A2J7PM83 A0A0T6B0U1 A0A1L8DJ27 A0A1L8DJ92 A0A1B6C415 A0A224XK28 A0A1Y1KWI5 A0A034VHS7 A0A2H4N0P7 A0A1W6EVS5 A0A1I8PLP7 A0A1B6EPA7 Q5QDM4 I5APT0 Q17EV1 A0A1B6L3K3 A0A1Q3G0V1 Q16926 A0A3B0K7F6 A0A1I8MU29 T1PJ76 A0A1B6H7M9 A0A2S2NKX1 A0A158P1F2 A0A2H8U075 A0A0P5N8V2 A0A0N8ASC2 A0A0P5GAN4 A0A0Q9WLP1 A0A0P5PCZ8 A0A0P5JB96 A0A0P6HCS7 A0A0P5LRL7 A0A0M4F2I2 A0A0P5GE64 A0A2M4BB49 A0A0R1E5W8 A0A2M4BBD6 A0A2M4BBB0 A0A2M4CUS9 A0A2M4CJJ1 A0A1I8JVB2 Q7PXC3 A0A0P5UCP7 Q1LZ40 A0A2M3Z5G8 A0A423ST97 A0A0A9ZBC7 V5IB95 V5H7R8 P30430 A0A293M166 A0A0P5YQS3 A0A0P5PKW8 A0A139WPK4 A0A0P8ZYW4 A0A088AAQ3 A0A1W4VK02 A0A293MYV8 E9GW88 A0A0K2UFU4 A0A0P5GJU5 A0A164LVR4 U4UME4 N6T225 A0A171A4N1 A0A0R3NK09 A0A0R3NJT9 A0A2T7NF47 B4GL59 A0A0P4WFA9 A0A1B6LWC5 A0A1B0GCW9
A0A194PYH4 A0A2J7PM83 A0A0T6B0U1 A0A1L8DJ27 A0A1L8DJ92 A0A1B6C415 A0A224XK28 A0A1Y1KWI5 A0A034VHS7 A0A2H4N0P7 A0A1W6EVS5 A0A1I8PLP7 A0A1B6EPA7 Q5QDM4 I5APT0 Q17EV1 A0A1B6L3K3 A0A1Q3G0V1 Q16926 A0A3B0K7F6 A0A1I8MU29 T1PJ76 A0A1B6H7M9 A0A2S2NKX1 A0A158P1F2 A0A2H8U075 A0A0P5N8V2 A0A0N8ASC2 A0A0P5GAN4 A0A0Q9WLP1 A0A0P5PCZ8 A0A0P5JB96 A0A0P6HCS7 A0A0P5LRL7 A0A0M4F2I2 A0A0P5GE64 A0A2M4BB49 A0A0R1E5W8 A0A2M4BBD6 A0A2M4BBB0 A0A2M4CUS9 A0A2M4CJJ1 A0A1I8JVB2 Q7PXC3 A0A0P5UCP7 Q1LZ40 A0A2M3Z5G8 A0A423ST97 A0A0A9ZBC7 V5IB95 V5H7R8 P30430 A0A293M166 A0A0P5YQS3 A0A0P5PKW8 A0A139WPK4 A0A0P8ZYW4 A0A088AAQ3 A0A1W4VK02 A0A293MYV8 E9GW88 A0A0K2UFU4 A0A0P5GJU5 A0A164LVR4 U4UME4 N6T225 A0A171A4N1 A0A0R3NK09 A0A0R3NJT9 A0A2T7NF47 B4GL59 A0A0P4WFA9 A0A1B6LWC5 A0A1B0GCW9
EC Number
3.4.21.75
Pubmed
EMBL
GDQN01002618
GDQN01002399
JAT88436.1
JAT88655.1
NWSH01000634
PCG75136.1
+ More
RSAL01000035 RVE51309.1 ODYU01012033 SOQ58118.1 PCG75135.1 AGBW02010967 OWR47458.1 KQ459585 KPI98386.1 NEVH01024418 PNF17448.1 LJIG01022461 KRT80455.1 GFDF01007653 JAV06431.1 GFDF01007654 JAV06430.1 GEDC01029050 JAS08248.1 GFTR01008057 JAW08369.1 GEZM01077633 JAV63227.1 GAKP01016893 JAC42059.1 KY653074 ATU47268.1 KY563396 ARK19805.1 GECZ01030000 GECZ01006849 JAS39769.1 JAS62920.1 AY682745 AAV74200.1 CM000070 EIM52965.2 CH477279 EAT45000.1 GEBQ01021727 JAT18250.1 GFDL01001616 JAV33429.1 L46373 AAC37262.1 OUUW01000008 SPP83990.1 KA648170 AFP62799.1 GECU01037000 JAS70706.1 GGMR01005168 MBY17787.1 ADTU01006445 ADTU01006446 GFXV01007606 MBW19411.1 GDIQ01145610 JAL06116.1 GDIQ01247683 JAK04042.1 GDIQ01243794 JAK07931.1 CH940650 KRF82871.1 GDIQ01206798 GDIQ01132577 GDIQ01117331 JAL19149.1 GDIQ01202199 GDIQ01069534 JAK49526.1 GDIQ01021367 JAN73370.1 GDIQ01166996 JAK84729.1 CP012526 ALC45295.1 GDIQ01243793 JAK07932.1 GGFJ01001070 MBW50211.1 CM000160 KRK04473.1 GGFJ01001202 MBW50343.1 GGFJ01001185 MBW50326.1 GGFL01004928 MBW69106.1 GGFL01001352 MBW65530.1 AAAB01008987 EAA01324.4 GDIP01114937 JAL88777.1 BT025186 ABF02288.1 GGFM01003009 MBW23760.1 QCYY01002810 ROT67420.1 GBHO01016547 GBHO01003116 GBHO01002963 GBHO01002961 JAG27057.1 JAG40488.1 JAG40641.1 JAG40643.1 GANP01015827 JAB68641.1 GANP01011359 JAB73109.1 M81431 L12370 L12369 L12372 AE014297 GFWV01009049 MAA33778.1 GDIP01054992 JAM48723.1 GDIQ01148322 JAL03404.1 KQ971307 KYB29761.1 CH902617 KPU79816.1 KPU79818.1 KPU79819.1 GFWV01020383 MAA45111.1 GL732569 EFX76306.1 HACA01019574 CDW36935.1 GDIQ01241425 JAK10300.1 LRGB01003123 KZS04479.1 KB632384 ERL94287.1 APGK01047137 APGK01047138 APGK01047139 KB741077 ENN74154.1 GEMB01001503 JAS01653.1 KRT01227.1 KRT01226.1 PZQS01000013 PVD19786.1 CH479185 EDW38283.1 GDRN01075266 JAI63082.1 GEBQ01011986 JAT27991.1 CCAG010002242 CCAG010002243
RSAL01000035 RVE51309.1 ODYU01012033 SOQ58118.1 PCG75135.1 AGBW02010967 OWR47458.1 KQ459585 KPI98386.1 NEVH01024418 PNF17448.1 LJIG01022461 KRT80455.1 GFDF01007653 JAV06431.1 GFDF01007654 JAV06430.1 GEDC01029050 JAS08248.1 GFTR01008057 JAW08369.1 GEZM01077633 JAV63227.1 GAKP01016893 JAC42059.1 KY653074 ATU47268.1 KY563396 ARK19805.1 GECZ01030000 GECZ01006849 JAS39769.1 JAS62920.1 AY682745 AAV74200.1 CM000070 EIM52965.2 CH477279 EAT45000.1 GEBQ01021727 JAT18250.1 GFDL01001616 JAV33429.1 L46373 AAC37262.1 OUUW01000008 SPP83990.1 KA648170 AFP62799.1 GECU01037000 JAS70706.1 GGMR01005168 MBY17787.1 ADTU01006445 ADTU01006446 GFXV01007606 MBW19411.1 GDIQ01145610 JAL06116.1 GDIQ01247683 JAK04042.1 GDIQ01243794 JAK07931.1 CH940650 KRF82871.1 GDIQ01206798 GDIQ01132577 GDIQ01117331 JAL19149.1 GDIQ01202199 GDIQ01069534 JAK49526.1 GDIQ01021367 JAN73370.1 GDIQ01166996 JAK84729.1 CP012526 ALC45295.1 GDIQ01243793 JAK07932.1 GGFJ01001070 MBW50211.1 CM000160 KRK04473.1 GGFJ01001202 MBW50343.1 GGFJ01001185 MBW50326.1 GGFL01004928 MBW69106.1 GGFL01001352 MBW65530.1 AAAB01008987 EAA01324.4 GDIP01114937 JAL88777.1 BT025186 ABF02288.1 GGFM01003009 MBW23760.1 QCYY01002810 ROT67420.1 GBHO01016547 GBHO01003116 GBHO01002963 GBHO01002961 JAG27057.1 JAG40488.1 JAG40641.1 JAG40643.1 GANP01015827 JAB68641.1 GANP01011359 JAB73109.1 M81431 L12370 L12369 L12372 AE014297 GFWV01009049 MAA33778.1 GDIP01054992 JAM48723.1 GDIQ01148322 JAL03404.1 KQ971307 KYB29761.1 CH902617 KPU79816.1 KPU79818.1 KPU79819.1 GFWV01020383 MAA45111.1 GL732569 EFX76306.1 HACA01019574 CDW36935.1 GDIQ01241425 JAK10300.1 LRGB01003123 KZS04479.1 KB632384 ERL94287.1 APGK01047137 APGK01047138 APGK01047139 KB741077 ENN74154.1 GEMB01001503 JAS01653.1 KRT01227.1 KRT01226.1 PZQS01000013 PVD19786.1 CH479185 EDW38283.1 GDRN01075266 JAI63082.1 GEBQ01011986 JAT27991.1 CCAG010002242 CCAG010002243
Proteomes
UP000218220
UP000283053
UP000007151
UP000053268
UP000235965
UP000095300
+ More
UP000001819 UP000008820 UP000268350 UP000095301 UP000005205 UP000008792 UP000092553 UP000002282 UP000075903 UP000007062 UP000283509 UP000000803 UP000007266 UP000007801 UP000005203 UP000192221 UP000000305 UP000076858 UP000030742 UP000019118 UP000245119 UP000008744 UP000092444
UP000001819 UP000008820 UP000268350 UP000095301 UP000005205 UP000008792 UP000092553 UP000002282 UP000075903 UP000007062 UP000283509 UP000000803 UP000007266 UP000007801 UP000005203 UP000192221 UP000000305 UP000076858 UP000030742 UP000019118 UP000245119 UP000008744 UP000092444
Interpro
IPR034182
Kexin/furin
+ More
IPR023828 Peptidase_S8_Ser-AS
IPR023827 Peptidase_S8_Asp-AS
IPR002884 P_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR022398 Peptidase_S8_His-AS
IPR008979 Galactose-bd-like_sf
IPR036852 Peptidase_S8/S53_dom_sf
IPR006212 Furin_repeat
IPR015500 Peptidase_S8_subtilisin-rel
IPR000209 Peptidase_S8/S53_dom
IPR038466 S8_pro-domain_sf
IPR032815 S8_pro-domain
IPR023828 Peptidase_S8_Ser-AS
IPR023827 Peptidase_S8_Asp-AS
IPR002884 P_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR022398 Peptidase_S8_His-AS
IPR008979 Galactose-bd-like_sf
IPR036852 Peptidase_S8/S53_dom_sf
IPR006212 Furin_repeat
IPR015500 Peptidase_S8_subtilisin-rel
IPR000209 Peptidase_S8/S53_dom
IPR038466 S8_pro-domain_sf
IPR032815 S8_pro-domain
Gene 3D
ProteinModelPortal
A0A1E1WNT8
A0A2A4JTN4
A0A437BLV5
A0A2H1WYJ9
A0A2A4JSW4
A0A212F132
+ More
A0A194PYH4 A0A2J7PM83 A0A0T6B0U1 A0A1L8DJ27 A0A1L8DJ92 A0A1B6C415 A0A224XK28 A0A1Y1KWI5 A0A034VHS7 A0A2H4N0P7 A0A1W6EVS5 A0A1I8PLP7 A0A1B6EPA7 Q5QDM4 I5APT0 Q17EV1 A0A1B6L3K3 A0A1Q3G0V1 Q16926 A0A3B0K7F6 A0A1I8MU29 T1PJ76 A0A1B6H7M9 A0A2S2NKX1 A0A158P1F2 A0A2H8U075 A0A0P5N8V2 A0A0N8ASC2 A0A0P5GAN4 A0A0Q9WLP1 A0A0P5PCZ8 A0A0P5JB96 A0A0P6HCS7 A0A0P5LRL7 A0A0M4F2I2 A0A0P5GE64 A0A2M4BB49 A0A0R1E5W8 A0A2M4BBD6 A0A2M4BBB0 A0A2M4CUS9 A0A2M4CJJ1 A0A1I8JVB2 Q7PXC3 A0A0P5UCP7 Q1LZ40 A0A2M3Z5G8 A0A423ST97 A0A0A9ZBC7 V5IB95 V5H7R8 P30430 A0A293M166 A0A0P5YQS3 A0A0P5PKW8 A0A139WPK4 A0A0P8ZYW4 A0A088AAQ3 A0A1W4VK02 A0A293MYV8 E9GW88 A0A0K2UFU4 A0A0P5GJU5 A0A164LVR4 U4UME4 N6T225 A0A171A4N1 A0A0R3NK09 A0A0R3NJT9 A0A2T7NF47 B4GL59 A0A0P4WFA9 A0A1B6LWC5 A0A1B0GCW9
A0A194PYH4 A0A2J7PM83 A0A0T6B0U1 A0A1L8DJ27 A0A1L8DJ92 A0A1B6C415 A0A224XK28 A0A1Y1KWI5 A0A034VHS7 A0A2H4N0P7 A0A1W6EVS5 A0A1I8PLP7 A0A1B6EPA7 Q5QDM4 I5APT0 Q17EV1 A0A1B6L3K3 A0A1Q3G0V1 Q16926 A0A3B0K7F6 A0A1I8MU29 T1PJ76 A0A1B6H7M9 A0A2S2NKX1 A0A158P1F2 A0A2H8U075 A0A0P5N8V2 A0A0N8ASC2 A0A0P5GAN4 A0A0Q9WLP1 A0A0P5PCZ8 A0A0P5JB96 A0A0P6HCS7 A0A0P5LRL7 A0A0M4F2I2 A0A0P5GE64 A0A2M4BB49 A0A0R1E5W8 A0A2M4BBD6 A0A2M4BBB0 A0A2M4CUS9 A0A2M4CJJ1 A0A1I8JVB2 Q7PXC3 A0A0P5UCP7 Q1LZ40 A0A2M3Z5G8 A0A423ST97 A0A0A9ZBC7 V5IB95 V5H7R8 P30430 A0A293M166 A0A0P5YQS3 A0A0P5PKW8 A0A139WPK4 A0A0P8ZYW4 A0A088AAQ3 A0A1W4VK02 A0A293MYV8 E9GW88 A0A0K2UFU4 A0A0P5GJU5 A0A164LVR4 U4UME4 N6T225 A0A171A4N1 A0A0R3NK09 A0A0R3NJT9 A0A2T7NF47 B4GL59 A0A0P4WFA9 A0A1B6LWC5 A0A1B0GCW9
PDB
1P8J
E-value=0.00212272,
Score=96
Ontologies
GO
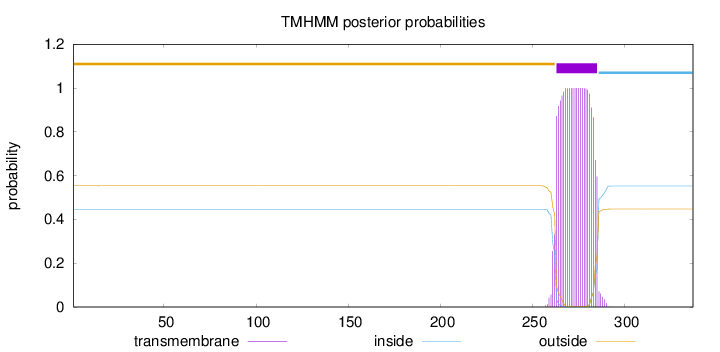
Topology
Subcellular location
Golgi apparatus membrane
Length:
337
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.61963
Exp number, first 60 AAs:
0.00071
Total prob of N-in:
0.44658
outside
1 - 262
TMhelix
263 - 285
inside
286 - 337
Population Genetic Test Statistics
Pi
217.477782
Theta
178.004845
Tajima's D
0.630843
CLR
0.006658
CSRT
0.551972401379931
Interpretation
Uncertain
