Gene
KWMTBOMO12523
Pre Gene Modal
BGIBMGA001512
Annotation
PREDICTED:_casein_kinase_I_isoform_gamma-3_isoform_X7_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 3.16
Sequence
CDS
ATGGTCGGCCCTAACTTTAGGGTCGGCAAGAAAATTGGTTGCGGCAACTTTGGTGAACTTAGACTCGGGAAAAATCTCTATAACAGCGACCATGTAGCGATCAAGATGGAGCCTATGAAGTCCAAGGCTCCGCAATTACATCTAGAATACAGATTTTACAAATTATTAGGGACGCACGAAGGTATACCGGAGGTGTATTACTTTGGTCCTTGCGGTAAATACAACGCTCTCGTAATGGAGTTGCTCGGGCCGTCGCTCGAGGACCTATTCGATCTATGCGGCCGTCGGTTCTCTCTAAAGACTGTGTTAATGATCGCTATGCAGTTGTTATACCGCATCGAGTATGTCCACACGAGACACCTAATATATCGCGACGTGAAGCCAGAGAATTTTTTAATAGGTAGAACTGCAACAAAAAAGGAGAAAATCGTGCACATCATTGATTTTGGTTTGGCCAAAGAATATATAGACTTAGAGACAAACAAACATATCCCATATCGGGAGCATAAGTCGCTCACAGGGACTGCTAGATACATGTCAATAAACACACATTTAGGAAAAGAACAATCGAGACGTGACGACCTAGAGGCTCTCGGACATATGTTTATGTATTTTTTACGTGGTTCCTTACCCTGGCAAGGTTTAAAAGCTGACACACTCAAAGAAAGATACCAAAAAATTGGCGACACTAAACGAGCGACCCCTATCGAGGTGCTATGCGAGGGCCACCCCGAGGAGCTGGCGACGTACCTGCGCTACGTGCGGCGGCTCGACTTCTTCGAGACCCCCGACTACGACCAGCTGCGAAGGATGTTCCGGGACCTGTTCGACAGGCGCGGATATATCGACGACGGAGAATTCGACTGGACCGGAAAGACAATGTCGACTCCTGTCGTGTCCGGCTCGACTGGTCAGGAAATCGTGATCTCACCGAACCGAGACAGTCACCAGCCGTTCAACAAGATGGGCACGTTGGCCGCCAAGGAGTTCGGCACAAGCGGCACGACTAATACCGGGAATTGGCCTCATAGCGGCAAGGCGACTCCGCTGACGACGGGGCATCTGACCCCCGCTGACAGGCACGGCTCCGTACAGGTGGTGAGCTCGACCAATGGCGAGCTTGGCGCCGACGACCCGACGGCGGGCCACAGCAACACGCCCATCACGGCGCCGCACCACGAGGTCGACGTGATTGACGACACCAAGTCAGTTTTCCAACTGTGCTGTTGTTTCTTCAAACGTAAAAAGAAGAAGTGCACCGCGCGCGCGGGCAAGTGA
Protein
MVGPNFRVGKKIGCGNFGELRLGKNLYNSDHVAIKMEPMKSKAPQLHLEYRFYKLLGTHEGIPEVYYFGPCGKYNALVMELLGPSLEDLFDLCGRRFSLKTVLMIAMQLLYRIEYVHTRHLIYRDVKPENFLIGRTATKKEKIVHIIDFGLAKEYIDLETNKHIPYREHKSLTGTARYMSINTHLGKEQSRRDDLEALGHMFMYFLRGSLPWQGLKADTLKERYQKIGDTKRATPIEVLCEGHPEELATYLRYVRRLDFFETPDYDQLRRMFRDLFDRRGYIDDGEFDWTGKTMSTPVVSGSTGQEIVISPNRDSHQPFNKMGTLAAKEFGTSGTTNTGNWPHSGKATPLTTGHLTPADRHGSVQVVSSTNGELGADDPTAGHSNTPITAPHHEVDVIDDTKSVFQLCCCFFKRKKKKCTARAGK
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
H9IW81
A0A2A4JMZ3
A0A3S2L607
A0A1Q3G276
Q17D24
Q17D25
+ More
A0A023F5N7 T1I5E2 A0A182J6E2 A0A1Q3G261 A0A069DTJ4 A0A0K8TPW6 A0A1L8DM43 A0A1B6E099 A0A1B6DM87 A0A195BF22 V5I8R0 A0A1Y9HEJ5 A0A2M3ZGA8 A0A2M4A6Y2 A0A1Y9GKZ2 A0A2M4A5H6 A0A1W4WE98 F5HM46 A0A1Y9HDN9 A0A1J1J416 A0A139WPM9 A0A158NBT7 A0A1Q3G285 A0A194Q4Q4 A0A2M4CSF6 D6W890 A0A1I8NRP5 A0A3B0JQQ8 A0A1W4WDX9 Q59DW8 A0A1W4W2D3 A0A026W5R2 B4JHB7 U4TYR5 A0A3B0JTX3 N6UKM0 I5APF1 Q9VEX2 A0A1Y9IVN6 A0A1Y1N112 A0A1B6LRJ7 A0A0A9XX81 A0A1Y1N7I7 A0A0A9ZEZ0 A0A146MCT2 A0A1Y9IUY2 A0A1B6J219 A0A1W4WFC0 A0A1I8NRN4 A0A3B0KFJ8 A0A2A3ETF9 A0A1W4WFC5 A0A1I8NRN7 K7IX80 A0A1Q3FUN8 A0A154NXA3 A0A1J1J424 A0A1Q3FV34 A0A232EJY1 A0A2M4CI64 A0A1B6MRR7 A0A0R3NPE9 A0A2M4AK71 A0A2M4CJA3 A0A2M3ZGB4 J9K738 A0A2M4CJK1 A0A2M4A5F8 A0A0A9WG97 A0A2H8TQ48 A0A1I8NRN5 A0A0A9WGA3 F5HM45 Q7Q272 A0A0K8V692 A0A0A1WLT6 A0A1L8DMU2 A0A2S2R1C3 B4LWT8 A0A2M4BMG9 B4K5S5 W8B908 A0A1Q3FUL9 A0A1I8NRN6 B3M2H7 A0A0B4KHL4 B4PR17 A0A0A1XRI6 A0A3B0KL25 A0A1W4W2C7 A0A3B0KHC9 A0A1Q3FUP5 Q86NK8 A0A1W4W2D5
A0A023F5N7 T1I5E2 A0A182J6E2 A0A1Q3G261 A0A069DTJ4 A0A0K8TPW6 A0A1L8DM43 A0A1B6E099 A0A1B6DM87 A0A195BF22 V5I8R0 A0A1Y9HEJ5 A0A2M3ZGA8 A0A2M4A6Y2 A0A1Y9GKZ2 A0A2M4A5H6 A0A1W4WE98 F5HM46 A0A1Y9HDN9 A0A1J1J416 A0A139WPM9 A0A158NBT7 A0A1Q3G285 A0A194Q4Q4 A0A2M4CSF6 D6W890 A0A1I8NRP5 A0A3B0JQQ8 A0A1W4WDX9 Q59DW8 A0A1W4W2D3 A0A026W5R2 B4JHB7 U4TYR5 A0A3B0JTX3 N6UKM0 I5APF1 Q9VEX2 A0A1Y9IVN6 A0A1Y1N112 A0A1B6LRJ7 A0A0A9XX81 A0A1Y1N7I7 A0A0A9ZEZ0 A0A146MCT2 A0A1Y9IUY2 A0A1B6J219 A0A1W4WFC0 A0A1I8NRN4 A0A3B0KFJ8 A0A2A3ETF9 A0A1W4WFC5 A0A1I8NRN7 K7IX80 A0A1Q3FUN8 A0A154NXA3 A0A1J1J424 A0A1Q3FV34 A0A232EJY1 A0A2M4CI64 A0A1B6MRR7 A0A0R3NPE9 A0A2M4AK71 A0A2M4CJA3 A0A2M3ZGB4 J9K738 A0A2M4CJK1 A0A2M4A5F8 A0A0A9WG97 A0A2H8TQ48 A0A1I8NRN5 A0A0A9WGA3 F5HM45 Q7Q272 A0A0K8V692 A0A0A1WLT6 A0A1L8DMU2 A0A2S2R1C3 B4LWT8 A0A2M4BMG9 B4K5S5 W8B908 A0A1Q3FUL9 A0A1I8NRN6 B3M2H7 A0A0B4KHL4 B4PR17 A0A0A1XRI6 A0A3B0KL25 A0A1W4W2C7 A0A3B0KHC9 A0A1Q3FUP5 Q86NK8 A0A1W4W2D5
Pubmed
19121390
17510324
25474469
26334808
26369729
12364791
+ More
14747013 17210077 18362917 19820115 21347285 26354079 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 17994087 23537049 15632085 28004739 25401762 26823975 20075255 28648823 25830018 24495485 17550304 24699734
14747013 17210077 18362917 19820115 21347285 26354079 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 17994087 23537049 15632085 28004739 25401762 26823975 20075255 28648823 25830018 24495485 17550304 24699734
EMBL
BABH01006020
BABH01006021
BABH01006022
NWSH01000993
PCG73189.1
RSAL01000128
+ More
RVE46495.1 GFDL01001143 JAV33902.1 CH477301 EAT44236.1 EAT44235.1 GBBI01002233 JAC16479.1 ACPB03003241 ACPB03003242 ACPB03003243 ACPB03003244 ACPB03003245 ACPB03003246 ACPB03003247 ACPB03003248 ACPB03003249 ACPB03003250 GFDL01001138 JAV33907.1 GBGD01001511 JAC87378.1 GDAI01001184 JAI16419.1 GFDF01006684 JAV07400.1 GEDC01005945 JAS31353.1 GEDC01010472 JAS26826.1 KQ976500 KYM83173.1 GALX01004250 JAB64216.1 GGFM01006772 MBW27523.1 GGFK01003181 MBW36502.1 APCN01002671 APCN01002672 APCN01002673 APCN01002674 APCN01002675 APCN01002676 APCN01002677 APCN01002678 GGFK01002708 MBW36029.1 AAAB01008978 EGK97357.1 CVRI01000070 CRL07157.1 KQ971307 KYB29873.1 ADTU01011318 ADTU01011319 ADTU01011320 ADTU01011321 ADTU01011322 ADTU01011323 GFDL01001131 JAV33914.1 KQ459585 KPI98390.1 GGFL01004065 MBW68243.1 EFA10912.2 OUUW01000008 SPP84507.1 AE014297 BT125862 AAO41569.1 ADU79250.1 KK107459 EZA50374.1 CH916369 EDV93824.1 KB631669 ERL85168.1 SPP84513.1 APGK01020129 KB740167 ENN81201.1 CM000070 EIM52836.2 AAF55293.1 GEZM01015512 GEZM01015510 GEZM01015509 JAV91579.1 GEBQ01025864 GEBQ01013691 JAT14113.1 JAT26286.1 GBHO01038089 GBHO01038088 GBHO01026981 GBHO01019608 JAG05515.1 JAG05516.1 JAG16623.1 JAG23996.1 GEZM01015514 GEZM01015513 GEZM01015511 GEZM01015508 JAV91577.1 GBHO01038085 GBHO01038082 GBHO01019612 GBHO01001113 JAG05519.1 JAG05522.1 JAG23992.1 JAG42491.1 GDHC01001018 JAQ17611.1 GECU01014494 JAS93212.1 SPP84506.1 KZ288192 PBC34331.1 AAZX01006551 GFDL01003748 JAV31297.1 KQ434778 KZC04305.1 CRL07155.1 GFDL01003752 JAV31293.1 NNAY01003912 OXU18667.1 GGFL01000862 MBW65040.1 GEBQ01031536 GEBQ01001369 JAT08441.1 JAT38608.1 KRT00880.1 GGFK01007850 MBW41171.1 GGFL01001151 MBW65329.1 GGFM01006774 MBW27525.1 ABLF02032388 GGFL01001362 MBW65540.1 GGFK01002702 MBW36023.1 GBHO01038087 GBHO01038086 GBHO01038084 GBHO01009559 JAG05517.1 JAG05518.1 JAG05520.1 JAG34045.1 GFXV01003573 MBW15378.1 GBHO01038081 GBHO01038080 GBHO01019618 GBHO01019609 JAG05523.1 JAG05524.1 JAG23986.1 JAG23995.1 EGK97356.1 EAA13659.5 GDHF01017860 JAI34454.1 GBXI01014646 JAC99645.1 GFDF01006400 JAV07684.1 GGMS01014624 MBY83827.1 CH940650 EDW66659.2 GGFJ01005136 MBW54277.1 CH933806 EDW15137.2 GAMC01020301 GAMC01020297 JAB86254.1 GFDL01003746 JAV31299.1 CH902617 EDV43430.2 AGB96004.1 CM000160 EDW97339.2 GBXI01001124 JAD13168.1 SPP84508.1 SPP84511.1 GFDL01003751 JAV31294.1 BT004871 AAO45227.1 ACZ94924.1
RVE46495.1 GFDL01001143 JAV33902.1 CH477301 EAT44236.1 EAT44235.1 GBBI01002233 JAC16479.1 ACPB03003241 ACPB03003242 ACPB03003243 ACPB03003244 ACPB03003245 ACPB03003246 ACPB03003247 ACPB03003248 ACPB03003249 ACPB03003250 GFDL01001138 JAV33907.1 GBGD01001511 JAC87378.1 GDAI01001184 JAI16419.1 GFDF01006684 JAV07400.1 GEDC01005945 JAS31353.1 GEDC01010472 JAS26826.1 KQ976500 KYM83173.1 GALX01004250 JAB64216.1 GGFM01006772 MBW27523.1 GGFK01003181 MBW36502.1 APCN01002671 APCN01002672 APCN01002673 APCN01002674 APCN01002675 APCN01002676 APCN01002677 APCN01002678 GGFK01002708 MBW36029.1 AAAB01008978 EGK97357.1 CVRI01000070 CRL07157.1 KQ971307 KYB29873.1 ADTU01011318 ADTU01011319 ADTU01011320 ADTU01011321 ADTU01011322 ADTU01011323 GFDL01001131 JAV33914.1 KQ459585 KPI98390.1 GGFL01004065 MBW68243.1 EFA10912.2 OUUW01000008 SPP84507.1 AE014297 BT125862 AAO41569.1 ADU79250.1 KK107459 EZA50374.1 CH916369 EDV93824.1 KB631669 ERL85168.1 SPP84513.1 APGK01020129 KB740167 ENN81201.1 CM000070 EIM52836.2 AAF55293.1 GEZM01015512 GEZM01015510 GEZM01015509 JAV91579.1 GEBQ01025864 GEBQ01013691 JAT14113.1 JAT26286.1 GBHO01038089 GBHO01038088 GBHO01026981 GBHO01019608 JAG05515.1 JAG05516.1 JAG16623.1 JAG23996.1 GEZM01015514 GEZM01015513 GEZM01015511 GEZM01015508 JAV91577.1 GBHO01038085 GBHO01038082 GBHO01019612 GBHO01001113 JAG05519.1 JAG05522.1 JAG23992.1 JAG42491.1 GDHC01001018 JAQ17611.1 GECU01014494 JAS93212.1 SPP84506.1 KZ288192 PBC34331.1 AAZX01006551 GFDL01003748 JAV31297.1 KQ434778 KZC04305.1 CRL07155.1 GFDL01003752 JAV31293.1 NNAY01003912 OXU18667.1 GGFL01000862 MBW65040.1 GEBQ01031536 GEBQ01001369 JAT08441.1 JAT38608.1 KRT00880.1 GGFK01007850 MBW41171.1 GGFL01001151 MBW65329.1 GGFM01006774 MBW27525.1 ABLF02032388 GGFL01001362 MBW65540.1 GGFK01002702 MBW36023.1 GBHO01038087 GBHO01038086 GBHO01038084 GBHO01009559 JAG05517.1 JAG05518.1 JAG05520.1 JAG34045.1 GFXV01003573 MBW15378.1 GBHO01038081 GBHO01038080 GBHO01019618 GBHO01019609 JAG05523.1 JAG05524.1 JAG23986.1 JAG23995.1 EGK97356.1 EAA13659.5 GDHF01017860 JAI34454.1 GBXI01014646 JAC99645.1 GFDF01006400 JAV07684.1 GGMS01014624 MBY83827.1 CH940650 EDW66659.2 GGFJ01005136 MBW54277.1 CH933806 EDW15137.2 GAMC01020301 GAMC01020297 JAB86254.1 GFDL01003746 JAV31299.1 CH902617 EDV43430.2 AGB96004.1 CM000160 EDW97339.2 GBXI01001124 JAD13168.1 SPP84508.1 SPP84511.1 GFDL01003751 JAV31294.1 BT004871 AAO45227.1 ACZ94924.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000008820
UP000015103
UP000075880
+ More
UP000078540 UP000075900 UP000075840 UP000192221 UP000007062 UP000183832 UP000007266 UP000005205 UP000053268 UP000095300 UP000268350 UP000000803 UP000053097 UP000001070 UP000030742 UP000019118 UP000001819 UP000075920 UP000192223 UP000242457 UP000002358 UP000076502 UP000215335 UP000007819 UP000008792 UP000009192 UP000007801 UP000002282
UP000078540 UP000075900 UP000075840 UP000192221 UP000007062 UP000183832 UP000007266 UP000005205 UP000053268 UP000095300 UP000268350 UP000000803 UP000053097 UP000001070 UP000030742 UP000019118 UP000001819 UP000075920 UP000192223 UP000242457 UP000002358 UP000076502 UP000215335 UP000007819 UP000008792 UP000009192 UP000007801 UP000002282
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9IW81
A0A2A4JMZ3
A0A3S2L607
A0A1Q3G276
Q17D24
Q17D25
+ More
A0A023F5N7 T1I5E2 A0A182J6E2 A0A1Q3G261 A0A069DTJ4 A0A0K8TPW6 A0A1L8DM43 A0A1B6E099 A0A1B6DM87 A0A195BF22 V5I8R0 A0A1Y9HEJ5 A0A2M3ZGA8 A0A2M4A6Y2 A0A1Y9GKZ2 A0A2M4A5H6 A0A1W4WE98 F5HM46 A0A1Y9HDN9 A0A1J1J416 A0A139WPM9 A0A158NBT7 A0A1Q3G285 A0A194Q4Q4 A0A2M4CSF6 D6W890 A0A1I8NRP5 A0A3B0JQQ8 A0A1W4WDX9 Q59DW8 A0A1W4W2D3 A0A026W5R2 B4JHB7 U4TYR5 A0A3B0JTX3 N6UKM0 I5APF1 Q9VEX2 A0A1Y9IVN6 A0A1Y1N112 A0A1B6LRJ7 A0A0A9XX81 A0A1Y1N7I7 A0A0A9ZEZ0 A0A146MCT2 A0A1Y9IUY2 A0A1B6J219 A0A1W4WFC0 A0A1I8NRN4 A0A3B0KFJ8 A0A2A3ETF9 A0A1W4WFC5 A0A1I8NRN7 K7IX80 A0A1Q3FUN8 A0A154NXA3 A0A1J1J424 A0A1Q3FV34 A0A232EJY1 A0A2M4CI64 A0A1B6MRR7 A0A0R3NPE9 A0A2M4AK71 A0A2M4CJA3 A0A2M3ZGB4 J9K738 A0A2M4CJK1 A0A2M4A5F8 A0A0A9WG97 A0A2H8TQ48 A0A1I8NRN5 A0A0A9WGA3 F5HM45 Q7Q272 A0A0K8V692 A0A0A1WLT6 A0A1L8DMU2 A0A2S2R1C3 B4LWT8 A0A2M4BMG9 B4K5S5 W8B908 A0A1Q3FUL9 A0A1I8NRN6 B3M2H7 A0A0B4KHL4 B4PR17 A0A0A1XRI6 A0A3B0KL25 A0A1W4W2C7 A0A3B0KHC9 A0A1Q3FUP5 Q86NK8 A0A1W4W2D5
A0A023F5N7 T1I5E2 A0A182J6E2 A0A1Q3G261 A0A069DTJ4 A0A0K8TPW6 A0A1L8DM43 A0A1B6E099 A0A1B6DM87 A0A195BF22 V5I8R0 A0A1Y9HEJ5 A0A2M3ZGA8 A0A2M4A6Y2 A0A1Y9GKZ2 A0A2M4A5H6 A0A1W4WE98 F5HM46 A0A1Y9HDN9 A0A1J1J416 A0A139WPM9 A0A158NBT7 A0A1Q3G285 A0A194Q4Q4 A0A2M4CSF6 D6W890 A0A1I8NRP5 A0A3B0JQQ8 A0A1W4WDX9 Q59DW8 A0A1W4W2D3 A0A026W5R2 B4JHB7 U4TYR5 A0A3B0JTX3 N6UKM0 I5APF1 Q9VEX2 A0A1Y9IVN6 A0A1Y1N112 A0A1B6LRJ7 A0A0A9XX81 A0A1Y1N7I7 A0A0A9ZEZ0 A0A146MCT2 A0A1Y9IUY2 A0A1B6J219 A0A1W4WFC0 A0A1I8NRN4 A0A3B0KFJ8 A0A2A3ETF9 A0A1W4WFC5 A0A1I8NRN7 K7IX80 A0A1Q3FUN8 A0A154NXA3 A0A1J1J424 A0A1Q3FV34 A0A232EJY1 A0A2M4CI64 A0A1B6MRR7 A0A0R3NPE9 A0A2M4AK71 A0A2M4CJA3 A0A2M3ZGB4 J9K738 A0A2M4CJK1 A0A2M4A5F8 A0A0A9WG97 A0A2H8TQ48 A0A1I8NRN5 A0A0A9WGA3 F5HM45 Q7Q272 A0A0K8V692 A0A0A1WLT6 A0A1L8DMU2 A0A2S2R1C3 B4LWT8 A0A2M4BMG9 B4K5S5 W8B908 A0A1Q3FUL9 A0A1I8NRN6 B3M2H7 A0A0B4KHL4 B4PR17 A0A0A1XRI6 A0A3B0KL25 A0A1W4W2C7 A0A3B0KHC9 A0A1Q3FUP5 Q86NK8 A0A1W4W2D5
PDB
4NT4
E-value=1.5652e-152,
Score=1384
Ontologies
KEGG
GO
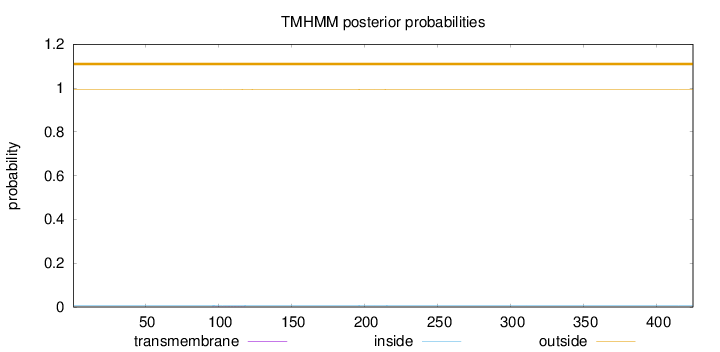
Topology
Length:
425
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01757
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.00648
outside
1 - 425
Population Genetic Test Statistics
Pi
192.584836
Theta
169.581558
Tajima's D
0.307871
CLR
0.613915
CSRT
0.450427478626069
Interpretation
Uncertain
