Gene
KWMTBOMO12507
Pre Gene Modal
BGIBMGA002389
Annotation
PREDICTED:_mucin-2-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.281
Sequence
CDS
ATGGTCCATGTCCCCAAAGCAAAGCGGACTAAATGGAACAAGAAGGCCAATGAACACATCCTGATTGGATACTCAGAGAGTACCAAAGGATACAGGCTGTACAATGTAAAAACAAAAAATATTACCACTAGCAGGGATGTTGTTATCATGGAGAAAATCAACGACACAGATATGATACAAGCAGTAGTGGAAGAAAAAGAAACTATTCCAAGTGAAGAAGGAATACAAAAATGTGCTGTGACAAGTGAAGATTCCACTTGTGACTCACTAAATAGCACATACATGACAGATGATGCTACATATGTACCAGATGATACCAGTACCACCAGCTCAGATGAATTCTGTGATAGTAAATCGAATCACAAGTCAGAAAATCTGCAGCCAATAATTAAGGTGGAAAGGAGGACTAGGAAGAAACCAGACTGGTATGGGTACAATAACATATGTCAGAGCAGTGAGGAGGTGGACGACAATCTTCTCACTTATGAGGATGTAATGAGAGGATCAGAACAAGAAGAATGGTCAAAAGCCATGAAAGAGGAAATACAATCTTTCCATCAGAATGATGCTTGGGAGTTAGTAGACAGACCCAGTGGTGAAAATATTGTGGCATGCAAATGGGTGTATAAAAAGAAACTTAACAGTGACAACTCAGTGAGATATAGGGCACGGAGTTAG
Protein
MVHVPKAKRTKWNKKANEHILIGYSESTKGYRLYNVKTKNITTSRDVVIMEKINDTDMIQAVVEEKETIPSEEGIQKCAVTSEDSTCDSLNSTYMTDDATYVPDDTSTTSSDEFCDSKSNHKSENLQPIIKVERRTRKKPDWYGYNNICQSSEEVDDNLLTYEDVMRGSEQEEWSKAMKEEIQSFHQNDAWELVDRPSGENIVACKWVYKKKLNSDNSVRYRARS
Summary
Uniprot
H9IYQ5
V5GAG2
A0A2A4J6E2
A0A1Y1NAZ7
A0A1Y1N668
A0A1Y1KL90
+ More
A0A182GPU0 A0A0A9XV68 A0A0A9XY84 A0A146LH62 A0A182H4F6 A0A194RJV9 A0A2M4CJW8 A0A034WM55 A0A2M4CK71 A0A1Y1N932 A0A1Y1N664 A0A1X7U495 T1PKV2 A0A182GLR8 A0A151SDY6 A0A151R2G6 A0A151RJN4 A0A151TKT1 A0A1W7R6A5 A0A182HAC3 A0A182H8E6 A0A151U026 A0A2J7RGN5 A0A2J7R937 A0A0J7NHJ8 A0A2J7R668 A0A2J7R8R7 A0A2J7Q1Z2 A0A2J7R8S8 A0A2J7RKJ6 A0A438E2Y0 A0A151R6U8 A0A2R6RIX9 J9L0N0 A0A0J7K416 A0A2J7PSB4 A0A438HI91 A0A146M1T4 A0A146LHR9 A0A0A9YB53 A0A0A9W6P7 A0A2K3NXK0 A0A1Y1MYP5 A0A2J7QBP6 A0A2N9G9X7 A0A151RIL1 A0A0A9WPH5 A0A0A9WRE3 A0A2R6RYD4 A0A1Y1MYL6 A0A182GFZ6 A0A2Z6NK18 A0A438IZF5 A0A1Y1MYL8 A0A2R6QHR4 A0A2N9GK87 A0A085N572 V5G0R8 A0A2N9H5C1 A0A0A9YRD9 A0A0J7KM48 A0A151SQX0 A5BQM4 A0A445IYF4 A0A0V0TKD8 A0A2M4CV35 A0A355BBC5 A0A1J3ENS7 A0A085NU43 A0A151T5R0 A0A438HJE1 A0A438E4N9 A0A0V0RWJ3 A0A0J7K6A2 Q6L3N8 A0A438E432 A0A0P6IU13 A0A085MVP5 A0A0V1NVX6 A0A1Y1N3X2 A0A0J7K761 A0A0J7K7N3 A0A0V1DG02 A0A438D664 A0A0L9T5R0 A0A438HG45 A0A438H8G8 A0A2A4JRI1 A0A0A9YYX8 A0A355BEE2
A0A182GPU0 A0A0A9XV68 A0A0A9XY84 A0A146LH62 A0A182H4F6 A0A194RJV9 A0A2M4CJW8 A0A034WM55 A0A2M4CK71 A0A1Y1N932 A0A1Y1N664 A0A1X7U495 T1PKV2 A0A182GLR8 A0A151SDY6 A0A151R2G6 A0A151RJN4 A0A151TKT1 A0A1W7R6A5 A0A182HAC3 A0A182H8E6 A0A151U026 A0A2J7RGN5 A0A2J7R937 A0A0J7NHJ8 A0A2J7R668 A0A2J7R8R7 A0A2J7Q1Z2 A0A2J7R8S8 A0A2J7RKJ6 A0A438E2Y0 A0A151R6U8 A0A2R6RIX9 J9L0N0 A0A0J7K416 A0A2J7PSB4 A0A438HI91 A0A146M1T4 A0A146LHR9 A0A0A9YB53 A0A0A9W6P7 A0A2K3NXK0 A0A1Y1MYP5 A0A2J7QBP6 A0A2N9G9X7 A0A151RIL1 A0A0A9WPH5 A0A0A9WRE3 A0A2R6RYD4 A0A1Y1MYL6 A0A182GFZ6 A0A2Z6NK18 A0A438IZF5 A0A1Y1MYL8 A0A2R6QHR4 A0A2N9GK87 A0A085N572 V5G0R8 A0A2N9H5C1 A0A0A9YRD9 A0A0J7KM48 A0A151SQX0 A5BQM4 A0A445IYF4 A0A0V0TKD8 A0A2M4CV35 A0A355BBC5 A0A1J3ENS7 A0A085NU43 A0A151T5R0 A0A438HJE1 A0A438E4N9 A0A0V0RWJ3 A0A0J7K6A2 Q6L3N8 A0A438E432 A0A0P6IU13 A0A085MVP5 A0A0V1NVX6 A0A1Y1N3X2 A0A0J7K761 A0A0J7K7N3 A0A0V1DG02 A0A438D664 A0A0L9T5R0 A0A438HG45 A0A438H8G8 A0A2A4JRI1 A0A0A9YYX8 A0A355BEE2
Pubmed
EMBL
BABH01011079
BABH01011080
GALX01001351
JAB67115.1
NWSH01002797
PCG67521.1
+ More
GEZM01011855 JAV93396.1 GEZM01011856 JAV93393.1 GEZM01082909 JAV61231.1 JXUM01079187 KQ563112 KXJ74446.1 GBHO01019780 JAG23824.1 GBHO01019781 JAG23823.1 GDHC01011760 JAQ06869.1 JXUM01025282 KQ560717 KXJ81160.1 KQ460297 KPJ16236.1 GGFL01001458 MBW65636.1 GAKP01004084 JAC54868.1 GGFL01001457 MBW65635.1 GEZM01011857 JAV93390.1 GEZM01011859 JAV93384.1 KA648745 AFP63374.1 JXUM01073804 KQ562795 KXJ75116.1 KQ483417 KYP53032.1 KQ484186 KYP36645.1 KQ483702 KYP42748.1 CM003607 KYP67664.1 GEHC01000963 JAV46682.1 JXUM01122375 KQ566597 KXJ69988.1 JXUM01029822 KQ560865 KXJ80573.1 CM003604 KYP72625.1 NEVH01003760 PNF39996.1 NEVH01006721 PNF37343.1 LBMM01004924 KMQ91985.1 NEVH01006980 PNF36330.1 PNF37231.1 NEVH01019375 PNF22597.1 PNF37230.1 NEVH01002725 PNF41351.1 QGNW01001416 RVW42052.1 KQ484038 KYP38095.1 NKQK01000005 PSS29970.1 ABLF02023757 ABLF02028466 ABLF02059696 LBMM01014433 KMQ85183.1 NEVH01021934 PNF19224.1 QGNW01000218 RVW84195.1 GDHC01004875 JAQ13754.1 GDHC01010945 JAQ07684.1 GBHO01016859 JAG26745.1 GBHO01043059 JAG00545.1 ASHM01002084 PNY07765.1 GEZM01017327 JAV90764.1 NEVH01016296 PNF26014.1 OIVN01001656 SPC96275.1 KQ483722 KYP42321.1 GBHO01033237 JAG10367.1 GBHO01033235 JAG10369.1 NKQK01000002 PSS35035.1 GEZM01017329 JAV90762.1 JXUM01060983 KQ562129 KXJ76624.1 DF974079 GAU44848.1 QGNW01000072 RVX02086.1 GEZM01017328 JAV90763.1 NKQK01000016 PSS08126.1 OIVN01002345 SPD02856.1 KL367553 KFD64618.1 GALX01004861 JAB63605.1 OIVN01002828 SPD06754.1 GBHO01008810 JAG34794.1 LBMM01005619 KMQ91367.1 CM003613 KYP57183.1 AM467700 CAN72600.1 QZWG01000009 RZB91070.1 JYDJ01000231 KRX39500.1 GGFL01004951 MBW69129.1 DNYX01000169 HBK83011.1 GEVK01019149 JAU33683.1 KL367475 KFD72989.1 CM003610 KYP62383.1 QGNW01000214 RVW84557.1 QGNW01001395 RVW42706.1 JYDL01000068 KRX18751.1 LBMM01013184 KMQ85774.1 AC149291 AAT38758.1 QGNW01001404 RVW42505.1 GDUN01000695 JAN95224.1 KL367628 KFD61291.1 JYDM01000091 KRZ88014.1 GEZM01013472 GEZM01013471 GEZM01013470 JAV92539.1 LBMM01012652 KMQ86074.1 LBMM01012480 KMQ86201.1 JYDI01000006 KRY60373.1 QGNW01001777 RVW30943.1 KQ258279 KOM25686.1 QGNW01000228 RVW83440.1 QGNW01000261 RVW80785.1 NWSH01000816 PCG74073.1 GBHO01007301 JAG36303.1 DNYX01000353 HBK84078.1
GEZM01011855 JAV93396.1 GEZM01011856 JAV93393.1 GEZM01082909 JAV61231.1 JXUM01079187 KQ563112 KXJ74446.1 GBHO01019780 JAG23824.1 GBHO01019781 JAG23823.1 GDHC01011760 JAQ06869.1 JXUM01025282 KQ560717 KXJ81160.1 KQ460297 KPJ16236.1 GGFL01001458 MBW65636.1 GAKP01004084 JAC54868.1 GGFL01001457 MBW65635.1 GEZM01011857 JAV93390.1 GEZM01011859 JAV93384.1 KA648745 AFP63374.1 JXUM01073804 KQ562795 KXJ75116.1 KQ483417 KYP53032.1 KQ484186 KYP36645.1 KQ483702 KYP42748.1 CM003607 KYP67664.1 GEHC01000963 JAV46682.1 JXUM01122375 KQ566597 KXJ69988.1 JXUM01029822 KQ560865 KXJ80573.1 CM003604 KYP72625.1 NEVH01003760 PNF39996.1 NEVH01006721 PNF37343.1 LBMM01004924 KMQ91985.1 NEVH01006980 PNF36330.1 PNF37231.1 NEVH01019375 PNF22597.1 PNF37230.1 NEVH01002725 PNF41351.1 QGNW01001416 RVW42052.1 KQ484038 KYP38095.1 NKQK01000005 PSS29970.1 ABLF02023757 ABLF02028466 ABLF02059696 LBMM01014433 KMQ85183.1 NEVH01021934 PNF19224.1 QGNW01000218 RVW84195.1 GDHC01004875 JAQ13754.1 GDHC01010945 JAQ07684.1 GBHO01016859 JAG26745.1 GBHO01043059 JAG00545.1 ASHM01002084 PNY07765.1 GEZM01017327 JAV90764.1 NEVH01016296 PNF26014.1 OIVN01001656 SPC96275.1 KQ483722 KYP42321.1 GBHO01033237 JAG10367.1 GBHO01033235 JAG10369.1 NKQK01000002 PSS35035.1 GEZM01017329 JAV90762.1 JXUM01060983 KQ562129 KXJ76624.1 DF974079 GAU44848.1 QGNW01000072 RVX02086.1 GEZM01017328 JAV90763.1 NKQK01000016 PSS08126.1 OIVN01002345 SPD02856.1 KL367553 KFD64618.1 GALX01004861 JAB63605.1 OIVN01002828 SPD06754.1 GBHO01008810 JAG34794.1 LBMM01005619 KMQ91367.1 CM003613 KYP57183.1 AM467700 CAN72600.1 QZWG01000009 RZB91070.1 JYDJ01000231 KRX39500.1 GGFL01004951 MBW69129.1 DNYX01000169 HBK83011.1 GEVK01019149 JAU33683.1 KL367475 KFD72989.1 CM003610 KYP62383.1 QGNW01000214 RVW84557.1 QGNW01001395 RVW42706.1 JYDL01000068 KRX18751.1 LBMM01013184 KMQ85774.1 AC149291 AAT38758.1 QGNW01001404 RVW42505.1 GDUN01000695 JAN95224.1 KL367628 KFD61291.1 JYDM01000091 KRZ88014.1 GEZM01013472 GEZM01013471 GEZM01013470 JAV92539.1 LBMM01012652 KMQ86074.1 LBMM01012480 KMQ86201.1 JYDI01000006 KRY60373.1 QGNW01001777 RVW30943.1 KQ258279 KOM25686.1 QGNW01000228 RVW83440.1 QGNW01000261 RVW80785.1 NWSH01000816 PCG74073.1 GBHO01007301 JAG36303.1 DNYX01000353 HBK84078.1
Proteomes
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9IYQ5
V5GAG2
A0A2A4J6E2
A0A1Y1NAZ7
A0A1Y1N668
A0A1Y1KL90
+ More
A0A182GPU0 A0A0A9XV68 A0A0A9XY84 A0A146LH62 A0A182H4F6 A0A194RJV9 A0A2M4CJW8 A0A034WM55 A0A2M4CK71 A0A1Y1N932 A0A1Y1N664 A0A1X7U495 T1PKV2 A0A182GLR8 A0A151SDY6 A0A151R2G6 A0A151RJN4 A0A151TKT1 A0A1W7R6A5 A0A182HAC3 A0A182H8E6 A0A151U026 A0A2J7RGN5 A0A2J7R937 A0A0J7NHJ8 A0A2J7R668 A0A2J7R8R7 A0A2J7Q1Z2 A0A2J7R8S8 A0A2J7RKJ6 A0A438E2Y0 A0A151R6U8 A0A2R6RIX9 J9L0N0 A0A0J7K416 A0A2J7PSB4 A0A438HI91 A0A146M1T4 A0A146LHR9 A0A0A9YB53 A0A0A9W6P7 A0A2K3NXK0 A0A1Y1MYP5 A0A2J7QBP6 A0A2N9G9X7 A0A151RIL1 A0A0A9WPH5 A0A0A9WRE3 A0A2R6RYD4 A0A1Y1MYL6 A0A182GFZ6 A0A2Z6NK18 A0A438IZF5 A0A1Y1MYL8 A0A2R6QHR4 A0A2N9GK87 A0A085N572 V5G0R8 A0A2N9H5C1 A0A0A9YRD9 A0A0J7KM48 A0A151SQX0 A5BQM4 A0A445IYF4 A0A0V0TKD8 A0A2M4CV35 A0A355BBC5 A0A1J3ENS7 A0A085NU43 A0A151T5R0 A0A438HJE1 A0A438E4N9 A0A0V0RWJ3 A0A0J7K6A2 Q6L3N8 A0A438E432 A0A0P6IU13 A0A085MVP5 A0A0V1NVX6 A0A1Y1N3X2 A0A0J7K761 A0A0J7K7N3 A0A0V1DG02 A0A438D664 A0A0L9T5R0 A0A438HG45 A0A438H8G8 A0A2A4JRI1 A0A0A9YYX8 A0A355BEE2
A0A182GPU0 A0A0A9XV68 A0A0A9XY84 A0A146LH62 A0A182H4F6 A0A194RJV9 A0A2M4CJW8 A0A034WM55 A0A2M4CK71 A0A1Y1N932 A0A1Y1N664 A0A1X7U495 T1PKV2 A0A182GLR8 A0A151SDY6 A0A151R2G6 A0A151RJN4 A0A151TKT1 A0A1W7R6A5 A0A182HAC3 A0A182H8E6 A0A151U026 A0A2J7RGN5 A0A2J7R937 A0A0J7NHJ8 A0A2J7R668 A0A2J7R8R7 A0A2J7Q1Z2 A0A2J7R8S8 A0A2J7RKJ6 A0A438E2Y0 A0A151R6U8 A0A2R6RIX9 J9L0N0 A0A0J7K416 A0A2J7PSB4 A0A438HI91 A0A146M1T4 A0A146LHR9 A0A0A9YB53 A0A0A9W6P7 A0A2K3NXK0 A0A1Y1MYP5 A0A2J7QBP6 A0A2N9G9X7 A0A151RIL1 A0A0A9WPH5 A0A0A9WRE3 A0A2R6RYD4 A0A1Y1MYL6 A0A182GFZ6 A0A2Z6NK18 A0A438IZF5 A0A1Y1MYL8 A0A2R6QHR4 A0A2N9GK87 A0A085N572 V5G0R8 A0A2N9H5C1 A0A0A9YRD9 A0A0J7KM48 A0A151SQX0 A5BQM4 A0A445IYF4 A0A0V0TKD8 A0A2M4CV35 A0A355BBC5 A0A1J3ENS7 A0A085NU43 A0A151T5R0 A0A438HJE1 A0A438E4N9 A0A0V0RWJ3 A0A0J7K6A2 Q6L3N8 A0A438E432 A0A0P6IU13 A0A085MVP5 A0A0V1NVX6 A0A1Y1N3X2 A0A0J7K761 A0A0J7K7N3 A0A0V1DG02 A0A438D664 A0A0L9T5R0 A0A438HG45 A0A438H8G8 A0A2A4JRI1 A0A0A9YYX8 A0A355BEE2
Ontologies
PANTHER
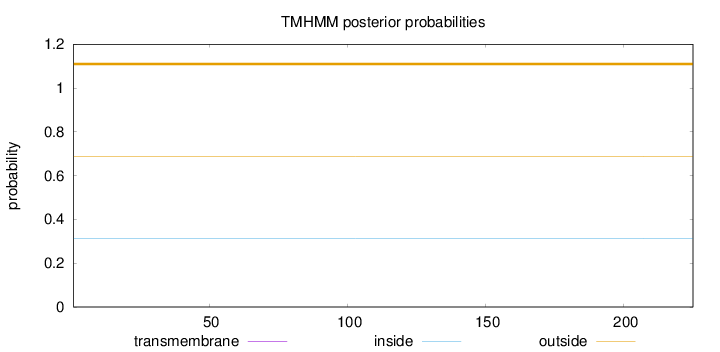
Topology
Length:
225
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0005
Exp number, first 60 AAs:
0.0005
Total prob of N-in:
0.31397
outside
1 - 225
Population Genetic Test Statistics
Pi
130.934303
Theta
100.298184
Tajima's D
0.957715
CLR
1.754203
CSRT
0.647217639118044
Interpretation
Uncertain
