Gene
KWMTBOMO12506
Pre Gene Modal
BGIBMGA009389
Annotation
PREDICTED:_mucin-2-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.049
Sequence
CDS
ATGGCTGGAAGTTACATTGTCAACGTGCCTAAATTGAAAGGTCGCGAAAACTTCGACGAGTGGGCGTTTGCAGCTGAACATTTTTTGATGTTGGAAGGTGTCGATATTCATAAGTTAGAATCGTCAACCGATGCTAGTTCTATTGATGAAAGAAAAGCTAAAGCTAAACTTATTATGACAATTGACTCTTCAATTTATGTTCACATTAAAAACGAGCAAACAGTACAAGGTGTGTGGAAGCGTTTAAAAGCCCTGTACGATGACTGTGGCTTTACGCGTCGCATCAGTCTACTTCGTCAGCTAATATCAATTCGATTAGAAGACAGCGAATCAATGGAAGCATATGTAACGCAAATGGTCGACACTGCACAAAAACTTAAAGGTACGGGTTTTCAGATCACTGATGAGTGGATCGGTTCACTATTATTAGCGGGCCTAACAGATACATTTGCTCCTATGATAATGGCCATTGAACATTCTGGATTAGCCATCAGTACGGATGCCATCAAGTCAAAATTACTGGACATGAGTTCAGAAGTTGGCAACACTGCAACTAACAATGCCTTTTTGTCGAAAGGTTGGCAGCACCGGAACAAGAAAAAAGGACTCAGGCGCAAGCTCACATATGATAACAGAAAAGAGTTGCTTACAGAATGTTTCCTATAA
Protein
MAGSYIVNVPKLKGRENFDEWAFAAEHFLMLEGVDIHKLESSTDASSIDERKAKAKLIMTIDSSIYVHIKNEQTVQGVWKRLKALYDDCGFTRRISLLRQLISIRLEDSESMEAYVTQMVDTAQKLKGTGFQITDEWIGSLLLAGLTDTFAPMIMAIEHSGLAISTDAIKSKLLDMSSEVGNTATNNAFLSKGWQHRNKKKGLRRKLTYDNRKELLTECFL
Summary
Uniprot
A0A2H1X3Q4
A0A2H1WQ59
A0A212F7V4
A0A2H1WW51
A0A1Y1N677
A0A1E1WHM3
+ More
A0A2H1V043 A0A1Y1N6L8 A0A1Y1N668 A0A1Y1N932 A0A2H1X258 A0A0A9Z2K2 A0A2A4JRI1 A0A3L8DS90 W4VRQ4 A0A0P6K155 A0A0J7K7C8 A0A1E1WH48 A0A2A4J959 A0A194RU38 A0A0J7JZV2 A0A182GZK6 A0A0A9X9I5 H9JND2 A0A1B6E9D0 A0A1Y1LWI3 A0A0J7K1U9 A0A3L8D826 A0A0A9W8L1 W4VRP9 A0A0J7NHJ8 W4VRP2 A0A2H1VGX3 A0A0A9YKS3 A0A0A9XCL0 A0A0J7MS03 A0A0A9XEY5 A0A224XHZ7 A0A0J7N4R7 A0A0A9Z9B6 A0A0A9ZER2 A0A0J7N3H6 A0A026WQ65 A0A194RJV9 A0A224XA26 A0A0J7KAH3 A0A0A1WXQ3 A0A0A9YWZ9 A0A026VU99 A0A0J7KK14 A0A0J7K9Y8 A0A0A9Y7E3 A0A034V012 A0A0A9XVJ9 A0A0A9WPH5 A0A0A9WRE3 A0A0A9W9K3 A0A0A9WBI3 A0A2A4K3A2 A0A0A9XZF1 A0A2W1BXU7 A0A437BGC2 A0A1Y1LX65 A0A0J7KCB5 A0A1I8NJG4 A0A1W7R6J6 A0A1W7R6K6 K7JXA3 A0A0J7N2D7 A0A0L7K8U3 A0A034VP70 A0A0L7LJJ1 A0A0J7K5G8 A0A1W7R670 A0A1W7R663 A0A2H1WMK4 A0A0A9YFM0 A0A3L8DXI8 A0A151I9Q8 A0A0A9WQD4
A0A2H1V043 A0A1Y1N6L8 A0A1Y1N668 A0A1Y1N932 A0A2H1X258 A0A0A9Z2K2 A0A2A4JRI1 A0A3L8DS90 W4VRQ4 A0A0P6K155 A0A0J7K7C8 A0A1E1WH48 A0A2A4J959 A0A194RU38 A0A0J7JZV2 A0A182GZK6 A0A0A9X9I5 H9JND2 A0A1B6E9D0 A0A1Y1LWI3 A0A0J7K1U9 A0A3L8D826 A0A0A9W8L1 W4VRP9 A0A0J7NHJ8 W4VRP2 A0A2H1VGX3 A0A0A9YKS3 A0A0A9XCL0 A0A0J7MS03 A0A0A9XEY5 A0A224XHZ7 A0A0J7N4R7 A0A0A9Z9B6 A0A0A9ZER2 A0A0J7N3H6 A0A026WQ65 A0A194RJV9 A0A224XA26 A0A0J7KAH3 A0A0A1WXQ3 A0A0A9YWZ9 A0A026VU99 A0A0J7KK14 A0A0J7K9Y8 A0A0A9Y7E3 A0A034V012 A0A0A9XVJ9 A0A0A9WPH5 A0A0A9WRE3 A0A0A9W9K3 A0A0A9WBI3 A0A2A4K3A2 A0A0A9XZF1 A0A2W1BXU7 A0A437BGC2 A0A1Y1LX65 A0A0J7KCB5 A0A1I8NJG4 A0A1W7R6J6 A0A1W7R6K6 K7JXA3 A0A0J7N2D7 A0A0L7K8U3 A0A034VP70 A0A0L7LJJ1 A0A0J7K5G8 A0A1W7R670 A0A1W7R663 A0A2H1WMK4 A0A0A9YFM0 A0A3L8DXI8 A0A151I9Q8 A0A0A9WQD4
Pubmed
EMBL
ODYU01013273
SOQ59980.1
ODYU01010175
SOQ55116.1
AGBW02009826
OWR49812.1
+ More
ODYU01011515 SOQ57290.1 GEZM01011859 GEZM01011855 JAV93394.1 GDQN01004560 JAT86494.1 ODYU01000075 SOQ34223.1 GEZM01011858 GEZM01011854 JAV93389.1 GEZM01011856 JAV93393.1 GEZM01011857 JAV93390.1 ODYU01012902 SOQ59440.1 GBHO01007564 JAG36040.1 NWSH01000816 PCG74073.1 QOIP01000004 RLU23300.1 GANO01000510 JAB59361.1 GDUN01000078 JAN95841.1 LBMM01012652 KMQ86076.1 GDQN01004734 JAT86320.1 NWSH01002292 PCG68657.1 PCG68658.1 KQ459875 KPJ19636.1 LBMM01019247 KMQ83577.1 JXUM01099713 KQ564513 KXJ72145.1 GBHO01029844 JAG13760.1 BABH01019436 GEDC01002834 JAS34464.1 GEZM01045162 JAV77889.1 LBMM01016836 KMQ84297.1 QOIP01000011 RLU16434.1 GBHO01042434 JAG01170.1 GANO01000659 JAB59212.1 LBMM01004924 KMQ91985.1 GANO01000660 JAB59211.1 ODYU01002507 SOQ40093.1 GBHO01011901 JAG31703.1 GBHO01026223 JAG17381.1 LBMM01020445 KMQ83285.1 GBHO01026221 JAG17383.1 GFTR01004319 JAW12107.1 LBMM01010129 KMQ87655.1 GBHO01001742 JAG41862.1 GBHO01001741 JAG41863.1 LBMM01010768 KMQ87255.1 KK107132 EZA58140.1 KQ460297 KPJ16236.1 GFTR01008662 JAW07764.1 LBMM01010791 KMQ87231.1 GBXI01011094 JAD03198.1 GBHO01007463 JAG36141.1 KK107871 EZA47378.1 LBMM01006235 KMQ90788.1 LBMM01010958 KMQ87117.1 GBHO01016596 JAG27008.1 GAKP01023107 JAC35849.1 GBHO01020706 JAG22898.1 GBHO01033237 JAG10367.1 GBHO01033235 JAG10369.1 GBHO01038507 JAG05097.1 GBHO01038505 JAG05099.1 NWSH01000234 PCG78150.1 GBHO01020974 JAG22630.1 KZ149913 PZC78057.1 RSAL01000063 RVE49487.1 GEZM01044588 JAV78134.1 LBMM01009660 KMQ87962.1 GEHC01000868 JAV46777.1 GEHC01000867 JAV46778.1 LBMM01011462 KMQ86820.1 JTDY01010240 KOB59525.1 GAKP01015352 JAC43600.1 JTDY01000892 KOB75534.1 LBMM01013295 KMQ85718.1 GEHC01000989 JAV46656.1 GEHC01000988 JAV46657.1 ODYU01009608 SOQ54162.1 GBHO01011746 GBHO01011744 GBHO01011742 JAG31858.1 JAG31860.1 JAG31862.1 QOIP01000003 RLU25147.1 KQ978269 KYM95809.1 GBHO01036544 JAG07060.1
ODYU01011515 SOQ57290.1 GEZM01011859 GEZM01011855 JAV93394.1 GDQN01004560 JAT86494.1 ODYU01000075 SOQ34223.1 GEZM01011858 GEZM01011854 JAV93389.1 GEZM01011856 JAV93393.1 GEZM01011857 JAV93390.1 ODYU01012902 SOQ59440.1 GBHO01007564 JAG36040.1 NWSH01000816 PCG74073.1 QOIP01000004 RLU23300.1 GANO01000510 JAB59361.1 GDUN01000078 JAN95841.1 LBMM01012652 KMQ86076.1 GDQN01004734 JAT86320.1 NWSH01002292 PCG68657.1 PCG68658.1 KQ459875 KPJ19636.1 LBMM01019247 KMQ83577.1 JXUM01099713 KQ564513 KXJ72145.1 GBHO01029844 JAG13760.1 BABH01019436 GEDC01002834 JAS34464.1 GEZM01045162 JAV77889.1 LBMM01016836 KMQ84297.1 QOIP01000011 RLU16434.1 GBHO01042434 JAG01170.1 GANO01000659 JAB59212.1 LBMM01004924 KMQ91985.1 GANO01000660 JAB59211.1 ODYU01002507 SOQ40093.1 GBHO01011901 JAG31703.1 GBHO01026223 JAG17381.1 LBMM01020445 KMQ83285.1 GBHO01026221 JAG17383.1 GFTR01004319 JAW12107.1 LBMM01010129 KMQ87655.1 GBHO01001742 JAG41862.1 GBHO01001741 JAG41863.1 LBMM01010768 KMQ87255.1 KK107132 EZA58140.1 KQ460297 KPJ16236.1 GFTR01008662 JAW07764.1 LBMM01010791 KMQ87231.1 GBXI01011094 JAD03198.1 GBHO01007463 JAG36141.1 KK107871 EZA47378.1 LBMM01006235 KMQ90788.1 LBMM01010958 KMQ87117.1 GBHO01016596 JAG27008.1 GAKP01023107 JAC35849.1 GBHO01020706 JAG22898.1 GBHO01033237 JAG10367.1 GBHO01033235 JAG10369.1 GBHO01038507 JAG05097.1 GBHO01038505 JAG05099.1 NWSH01000234 PCG78150.1 GBHO01020974 JAG22630.1 KZ149913 PZC78057.1 RSAL01000063 RVE49487.1 GEZM01044588 JAV78134.1 LBMM01009660 KMQ87962.1 GEHC01000868 JAV46777.1 GEHC01000867 JAV46778.1 LBMM01011462 KMQ86820.1 JTDY01010240 KOB59525.1 GAKP01015352 JAC43600.1 JTDY01000892 KOB75534.1 LBMM01013295 KMQ85718.1 GEHC01000989 JAV46656.1 GEHC01000988 JAV46657.1 ODYU01009608 SOQ54162.1 GBHO01011746 GBHO01011744 GBHO01011742 JAG31858.1 JAG31860.1 JAG31862.1 QOIP01000003 RLU25147.1 KQ978269 KYM95809.1 GBHO01036544 JAG07060.1
Proteomes
PRIDE
Pfam
Interpro
IPR001878
Znf_CCHC
+ More
IPR036875 Znf_CCHC_sf
IPR025724 GAG-pre-integrase_dom
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR039537 Retrotran_Ty1/copia-like
IPR012337 RNaseH-like_sf
IPR013103 RVT_2
IPR025314 DUF4219
IPR032049 Msl2-CXC
IPR033467 Tesmin/TSO1-like_CXC
IPR037922 MSL2
IPR041426 Mos1_HTH
IPR036875 Znf_CCHC_sf
IPR025724 GAG-pre-integrase_dom
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR039537 Retrotran_Ty1/copia-like
IPR012337 RNaseH-like_sf
IPR013103 RVT_2
IPR025314 DUF4219
IPR032049 Msl2-CXC
IPR033467 Tesmin/TSO1-like_CXC
IPR037922 MSL2
IPR041426 Mos1_HTH
Gene 3D
CDD
ProteinModelPortal
A0A2H1X3Q4
A0A2H1WQ59
A0A212F7V4
A0A2H1WW51
A0A1Y1N677
A0A1E1WHM3
+ More
A0A2H1V043 A0A1Y1N6L8 A0A1Y1N668 A0A1Y1N932 A0A2H1X258 A0A0A9Z2K2 A0A2A4JRI1 A0A3L8DS90 W4VRQ4 A0A0P6K155 A0A0J7K7C8 A0A1E1WH48 A0A2A4J959 A0A194RU38 A0A0J7JZV2 A0A182GZK6 A0A0A9X9I5 H9JND2 A0A1B6E9D0 A0A1Y1LWI3 A0A0J7K1U9 A0A3L8D826 A0A0A9W8L1 W4VRP9 A0A0J7NHJ8 W4VRP2 A0A2H1VGX3 A0A0A9YKS3 A0A0A9XCL0 A0A0J7MS03 A0A0A9XEY5 A0A224XHZ7 A0A0J7N4R7 A0A0A9Z9B6 A0A0A9ZER2 A0A0J7N3H6 A0A026WQ65 A0A194RJV9 A0A224XA26 A0A0J7KAH3 A0A0A1WXQ3 A0A0A9YWZ9 A0A026VU99 A0A0J7KK14 A0A0J7K9Y8 A0A0A9Y7E3 A0A034V012 A0A0A9XVJ9 A0A0A9WPH5 A0A0A9WRE3 A0A0A9W9K3 A0A0A9WBI3 A0A2A4K3A2 A0A0A9XZF1 A0A2W1BXU7 A0A437BGC2 A0A1Y1LX65 A0A0J7KCB5 A0A1I8NJG4 A0A1W7R6J6 A0A1W7R6K6 K7JXA3 A0A0J7N2D7 A0A0L7K8U3 A0A034VP70 A0A0L7LJJ1 A0A0J7K5G8 A0A1W7R670 A0A1W7R663 A0A2H1WMK4 A0A0A9YFM0 A0A3L8DXI8 A0A151I9Q8 A0A0A9WQD4
A0A2H1V043 A0A1Y1N6L8 A0A1Y1N668 A0A1Y1N932 A0A2H1X258 A0A0A9Z2K2 A0A2A4JRI1 A0A3L8DS90 W4VRQ4 A0A0P6K155 A0A0J7K7C8 A0A1E1WH48 A0A2A4J959 A0A194RU38 A0A0J7JZV2 A0A182GZK6 A0A0A9X9I5 H9JND2 A0A1B6E9D0 A0A1Y1LWI3 A0A0J7K1U9 A0A3L8D826 A0A0A9W8L1 W4VRP9 A0A0J7NHJ8 W4VRP2 A0A2H1VGX3 A0A0A9YKS3 A0A0A9XCL0 A0A0J7MS03 A0A0A9XEY5 A0A224XHZ7 A0A0J7N4R7 A0A0A9Z9B6 A0A0A9ZER2 A0A0J7N3H6 A0A026WQ65 A0A194RJV9 A0A224XA26 A0A0J7KAH3 A0A0A1WXQ3 A0A0A9YWZ9 A0A026VU99 A0A0J7KK14 A0A0J7K9Y8 A0A0A9Y7E3 A0A034V012 A0A0A9XVJ9 A0A0A9WPH5 A0A0A9WRE3 A0A0A9W9K3 A0A0A9WBI3 A0A2A4K3A2 A0A0A9XZF1 A0A2W1BXU7 A0A437BGC2 A0A1Y1LX65 A0A0J7KCB5 A0A1I8NJG4 A0A1W7R6J6 A0A1W7R6K6 K7JXA3 A0A0J7N2D7 A0A0L7K8U3 A0A034VP70 A0A0L7LJJ1 A0A0J7K5G8 A0A1W7R670 A0A1W7R663 A0A2H1WMK4 A0A0A9YFM0 A0A3L8DXI8 A0A151I9Q8 A0A0A9WQD4
Ontologies
KEGG
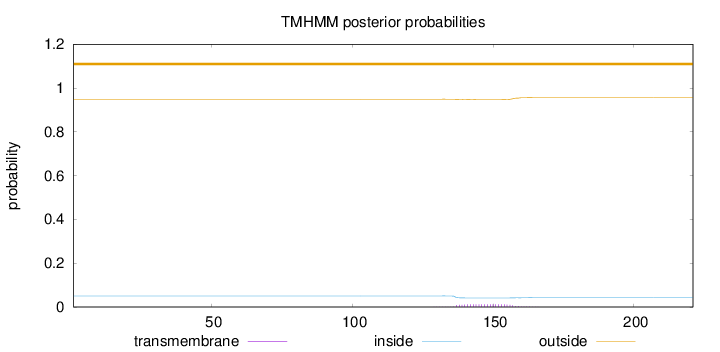
Topology
Length:
221
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2592
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.05072
outside
1 - 221
Population Genetic Test Statistics
Pi
47.417552
Theta
55.64954
Tajima's D
-0.415899
CLR
3.289916
CSRT
0.263986800659967
Interpretation
Uncertain
