Gene
KWMTBOMO12505
Pre Gene Modal
BGIBMGA001504
Annotation
peritrophin_1_[Mamestra_configurata]
Location in the cell
Extracellular Reliability : 1.801 Nuclear Reliability : 1.785
Sequence
CDS
ATGGAAAAATTTAAAGGTATTCTATTAGTATTATACGCAGTCGCATTGTCAAATGCATCTGTCATCAAAGAAAACACCAATAAAGCTACCAAAGGGGTCAATTTCGAATCCGGTAAAGCTACTGAAATCTGTGCTCGTATAGGTTCAGATGGTATACTAGTTGCTCATGAACATTGCACAAGATTTTATAAATGTGCAGAAGGTCGTCCTGTAGCACTTAAGTGCCCTCCAAATCTATTGTTTAATCCTAGCAACGAGCAATGTGACTGGCCTCATAATGTAGAGTGTGGAGACAGAACCATTCCAGAAGACGACGATGATGTTGATAGTGAAAGTGACAATGGCAGTGGAGGAAACGGAAACGAAAACGATAACACTGGAAATGGACACGCCGACCCCAGCCTGGCTACTGAAATATGTGCTGAAAAAGATTCAGACGGTGTTCTGGTCGCCCATGAGCACTGCACCAGATTTTACAAATGTTTCGATAGTCATCCAGTAGCCCTCATTTGTCCTCCAAATCTGTTGTATAATCCTAATAACGAGCAGTGCGACTGGCCTCATAATGTAGAGTGTGGAGACAGAATCATTCCAGAAGACGACGAGGATGTTGATAGTGAAAGTGACAATGGCAGTGGAGGAAACGGAAACGAAAACGATAACACTGGAAATGGACACGCCGACCCCAGCCTGGCTACTGAAATATGTGCTGAAAAAGATTCAGACGGTGTTCTGGTCGCCCATGAGCACTGCACCAGATTTTACAAATGTTTCGATAGTCATCCAGTAGCCCTCATTTGTCCTCCAAATCTGTTGTATAATCCTAATAACGAGCAGTGCGACTGGCCTCATAATGTAGAGTGTGGAGACAGAACCATTCCAGAAGACGACGAGGATGTTGATAGTGAAAGTGACAATGGTAATGGAGGAAACGGAAACGAAAACGATAACACTGGAAATGGACACGCCGACCCCAGCCTGGCTACTGAAATATGTGCTGAAAAAGATTCAGACGGTGTTTTGGTCGCCCATGAGCACTGCACCAGATTTTACAAATGTTTCAATAGTCATCCAGTAGCCCTCATTTGTCCTCCAAATCTATTGTATAATCCTAATAACGAGCAGTGCGACTGGCCTCATAATGTAGAGTGTGGAGACAGAACCATTCCAGAAGACGACGAGGATGTTGATAGTGAAAGTGACAATGGCAGTGGAGGAAACGGAAACGAAAACGATAACACTGGAAATGGACACGCCGACCCCAGCCTGGCTACTGAAATATGTGCTGAAAAAGATTCAGACGGTGTTCTGGTCGCCCATGAGCACTGCACCAGATTTTACAAATGTTTCGATAGTCATCCAGTAGCCCTCATTTGTCCTCCAAATCTGTTGTATAATCCTAATAACGAGCAGTGCGACTGGCCTCATAATGTAGAGTGTGGAGACAGAACCATTCCAGAAGACGACGAGGATGTTGATAGTGAAAGTGACAATGGCAGTGGAGGAAACGGAAACGAAAACGATAACACTGGAAATGGACACGCCGACCCCAGCCTGGCTACTGAAATATGTGCTGAAAAAGATTCAGACGGTGTTCTGGTCGCCCATGAGCACTGCACCAGATTTTACAAATGTTTCGATAGTCATCCAGTAGCCCTCATTTGTCCTCCAAATCTGTTGTATAATCCTAATAACGAGCAGTGCGACTGGCCTCATAATGTAGAGTGTGGAGACAGAACCATTCCAGAAGACGACGAGGATGTTGATAGTGAAAGTGACAATGGTAATGGAGGAAACGGAAACGAAAACGATAACACTGGAAATGGACACGCTGACCCCAGCCTGGCTACTGAAATTTGTGCTGAAAATGATTCAGACGGTGTTTTGGTCGCCCATGAGCACTGCACCAGATTTTACAAATGTTTCAATAGTCGTCCAGTAGCCCTCATTTGTCCGCCAAATCTGTTGTATAATCCTAGTAACGAGCAGTGTGACTGGCCTCATAATGTAGAGTGTGGAGACAGAACCATTCCAGAAAACGTCGAGGATGAGGACAACGATAATGCCAGCGATGGCAATTTGGACGGAAACGAAAATGAAGGCAATGACAACAGCAGCGGTAACTGCGATCCCAGCGCAGCTCCTACGTTGTGTGCTCAAACTGGTTCAGATAGTGTTCTAATTGCCCACGAAAATTGTGATAAATTTTACATATGCTGGAACCACAAGCCTGTAGTTCTCAGCTGTCCTCCAAATCTCCTCTACAATCCATCGAAAGAAGAATGCGATTGGCCTCAGAATGTCGACTGCGGTTCAAGAATTATTCCATAA
Protein
MEKFKGILLVLYAVALSNASVIKENTNKATKGVNFESGKATEICARIGSDGILVAHEHCTRFYKCAEGRPVALKCPPNLLFNPSNEQCDWPHNVECGDRTIPEDDDDVDSESDNGSGGNGNENDNTGNGHADPSLATEICAEKDSDGVLVAHEHCTRFYKCFDSHPVALICPPNLLYNPNNEQCDWPHNVECGDRIIPEDDEDVDSESDNGSGGNGNENDNTGNGHADPSLATEICAEKDSDGVLVAHEHCTRFYKCFDSHPVALICPPNLLYNPNNEQCDWPHNVECGDRTIPEDDEDVDSESDNGNGGNGNENDNTGNGHADPSLATEICAEKDSDGVLVAHEHCTRFYKCFNSHPVALICPPNLLYNPNNEQCDWPHNVECGDRTIPEDDEDVDSESDNGSGGNGNENDNTGNGHADPSLATEICAEKDSDGVLVAHEHCTRFYKCFDSHPVALICPPNLLYNPNNEQCDWPHNVECGDRTIPEDDEDVDSESDNGSGGNGNENDNTGNGHADPSLATEICAEKDSDGVLVAHEHCTRFYKCFDSHPVALICPPNLLYNPNNEQCDWPHNVECGDRTIPEDDEDVDSESDNGNGGNGNENDNTGNGHADPSLATEICAENDSDGVLVAHEHCTRFYKCFNSRPVALICPPNLLYNPSNEQCDWPHNVECGDRTIPENVEDEDNDNASDGNLDGNENEGNDNSSGNCDPSAAPTLCAQTGSDSVLIAHENCDKFYICWNHKPVVLSCPPNLLYNPSKEECDWPQNVDCGSRIIP
Summary
Uniprot
Q86BV0
A0A2A4JPM5
Q6VAP0
B6CAP5
C4N8D2
A0A3S2M5L1
+ More
A0A212EJK6 A0A3S2LQ37 A0A2A4JAZ3 A0A194Q044 A0A2H1V8J7 A0A3R7PK84 C4N881 M1EYJ2 A0A1W5C8V7 D1MAJ8 A0A3G5BIJ9 M1EWR5 A0A1S4HEA2 E2C958 A0A088BU36 A0A151WLI9 A0A158N966 A0A195CTW7 A0A195FP78 A0A3S2NBP7 A0A195B4N7 A0A087T425 A0A3L8E3H8 A0A3L8E266
A0A212EJK6 A0A3S2LQ37 A0A2A4JAZ3 A0A194Q044 A0A2H1V8J7 A0A3R7PK84 C4N881 M1EYJ2 A0A1W5C8V7 D1MAJ8 A0A3G5BIJ9 M1EWR5 A0A1S4HEA2 E2C958 A0A088BU36 A0A151WLI9 A0A158N966 A0A195CTW7 A0A195FP78 A0A3S2NBP7 A0A195B4N7 A0A087T425 A0A3L8E3H8 A0A3L8E266
Pubmed
EMBL
AY277403
AAP33177.1
NWSH01000891
PCG73658.1
AY345124
AAR06265.1
+ More
EU139126 ABW98673.1 FJ408730 ACQ65651.1 RSAL01000033 RVE51508.1 AGBW02014451 OWR41650.1 RVE51509.1 NWSH01002232 PCG68868.1 KQ459585 KPI98364.1 ODYU01001235 SOQ37170.1 QCYY01001877 ROT74564.1 FJ393548 ACR20468.1 JF681185 AFD28281.1 AAAB01008986 EAA00508.4 GU128106 KQ971357 ACY95489.1 KYB26070.1 MK075202 AYV99605.1 JF681186 AFD28282.1 GL453780 EFN75531.1 KC703970 AHH02588.1 KQ982959 KYQ48703.1 ADTU01008736 ADTU01008737 KQ977279 KYN04138.1 KQ981382 KYN42258.1 RSAL01000265 RVE43282.1 KQ976618 KYM79154.1 KK113308 KFM59864.1 QOIP01000001 RLU27300.1 RLU26612.1
EU139126 ABW98673.1 FJ408730 ACQ65651.1 RSAL01000033 RVE51508.1 AGBW02014451 OWR41650.1 RVE51509.1 NWSH01002232 PCG68868.1 KQ459585 KPI98364.1 ODYU01001235 SOQ37170.1 QCYY01001877 ROT74564.1 FJ393548 ACR20468.1 JF681185 AFD28281.1 AAAB01008986 EAA00508.4 GU128106 KQ971357 ACY95489.1 KYB26070.1 MK075202 AYV99605.1 JF681186 AFD28282.1 GL453780 EFN75531.1 KC703970 AHH02588.1 KQ982959 KYQ48703.1 ADTU01008736 ADTU01008737 KQ977279 KYN04138.1 KQ981382 KYN42258.1 RSAL01000265 RVE43282.1 KQ976618 KYM79154.1 KK113308 KFM59864.1 QOIP01000001 RLU27300.1 RLU26612.1
Proteomes
Pfam
PF01607 CBM_14
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
Q86BV0
A0A2A4JPM5
Q6VAP0
B6CAP5
C4N8D2
A0A3S2M5L1
+ More
A0A212EJK6 A0A3S2LQ37 A0A2A4JAZ3 A0A194Q044 A0A2H1V8J7 A0A3R7PK84 C4N881 M1EYJ2 A0A1W5C8V7 D1MAJ8 A0A3G5BIJ9 M1EWR5 A0A1S4HEA2 E2C958 A0A088BU36 A0A151WLI9 A0A158N966 A0A195CTW7 A0A195FP78 A0A3S2NBP7 A0A195B4N7 A0A087T425 A0A3L8E3H8 A0A3L8E266
A0A212EJK6 A0A3S2LQ37 A0A2A4JAZ3 A0A194Q044 A0A2H1V8J7 A0A3R7PK84 C4N881 M1EYJ2 A0A1W5C8V7 D1MAJ8 A0A3G5BIJ9 M1EWR5 A0A1S4HEA2 E2C958 A0A088BU36 A0A151WLI9 A0A158N966 A0A195CTW7 A0A195FP78 A0A3S2NBP7 A0A195B4N7 A0A087T425 A0A3L8E3H8 A0A3L8E266
PDB
6G9E
E-value=7.42652e-10,
Score=156
Ontologies
GO
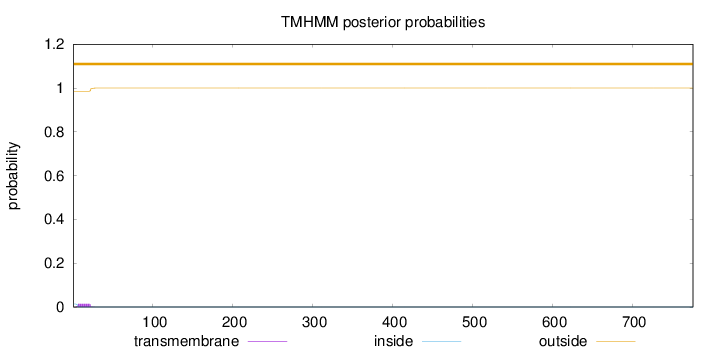
Topology
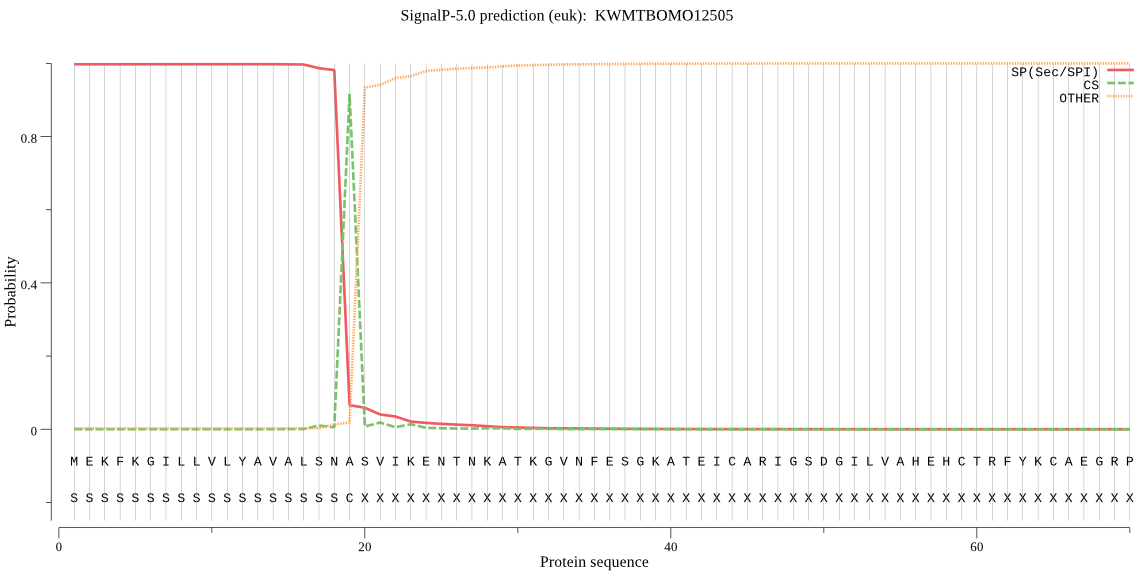
SignalP
Position: 1 - 19,
Likelihood: 0.997593
Length:
776
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23961
Exp number, first 60 AAs:
0.23961
Total prob of N-in:
0.01416
outside
1 - 776
Population Genetic Test Statistics
Pi
236.754828
Theta
150.354117
Tajima's D
2.096021
CLR
1.070455
CSRT
0.898155092245388
Interpretation
Uncertain
