Gene
KWMTBOMO12503
Annotation
PREDICTED:_lachesin-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.869
Sequence
CDS
ATGCGGCACCAGACTACATCTCCACTAGGCCGGGTGGGACCCACTGCGTCCCTGCGAGACTCCGAGCCCGACTTTGCTGGACCAATAGAAAACGTCACAGTAGCTCTCGGCAGGGAAGCCATACTCACCTGCTCAGTCACCGACCTTGGAGACTACAAGGTAGCCTGGATTCGAGCAGATGATCAAACTATCCTCACACTCCACACGAGGCTGGTGACGCACAGCTCTCGGTATGCGGTTACCAACGATTCTCCGCATTCCTGGCAACTCCACATAAGACCTCTGAAGGTAGAAGATCGGGGCTGCTACATGTGCCAGATAAATACCAGCACGATGAAGAAGCAGATCGGCTGTGTGAATGTGCTAGTTCCACCGAACATCGTAGATAAAGGCACGAGTGGAGATCTCGTTGCACGCGAAGGTCAAGACACAAGTCTATCCTGCAAAGCCGAAGGAACACCACTCCCGCGAATTTTGTGGAGAAGAGAAGACGGTACCAACATTCAACTCAGGAATGAAGCCGGCGATCTTCGAAAAGTTGACATGTTCATGGGACCAAATTTAAACCTAACAAAAGTAGAACGACGACAGATGGGAGCATACCTTTGTATCGCGTCAAACGAAGTTCCACCTTCAGTCAGCAAAAGGATAATGCTGAATGTTAATTTTCCACCATCAATCCTGGTGGCGACCAAGGTCATCGGCGTGCCGACAGGATCACAAGCCAAACTGGAATGTTTCGTCGAGGCGTTTCCACAAGCTATTAATTATTGGCTCAAGAGCGGCGAAGAGATGATTCTTTCCAATTCCAAATATAATTTAACGGAAGATCGAATATCCCAGTACCAACTCCGAACCATTCTACAGATTTCGAAGTTTGAGCCTGAAGATATAGGCACCTACACGTGCGTCTCCACAAATACGATGGGAAAGTCGGAAGGGACATTAAGACTTTATGGTGAGTTTTTATGA
Protein
MRHQTTSPLGRVGPTASLRDSEPDFAGPIENVTVALGREAILTCSVTDLGDYKVAWIRADDQTILTLHTRLVTHSSRYAVTNDSPHSWQLHIRPLKVEDRGCYMCQINTSTMKKQIGCVNVLVPPNIVDKGTSGDLVAREGQDTSLSCKAEGTPLPRILWRREDGTNIQLRNEAGDLRKVDMFMGPNLNLTKVERRQMGAYLCIASNEVPPSVSKRIMLNVNFPPSILVATKVIGVPTGSQAKLECFVEAFPQAINYWLKSGEEMILSNSKYNLTEDRISQYQLRTILQISKFEPEDIGTYTCVSTNTMGKSEGTLRLYGEFL
Summary
Uniprot
A0A3S2NFW5
A0A212F862
A0A2A4IVJ7
A0A194Q082
A0A0N0PCD5
D6WCY6
+ More
A0A1B6DBF6 A0A1B6KXD0 A0A139WNB8 E0VYZ0 D6WCY9 A0A139WN83 A0A1W4XLM5 A0A2A4K566 A0A437B3S3 A0A194QC96 A0A194RDC2 A0A1B0GE93 A0A067RKA0 A0A1A9Z6V7 A0A1A9XQ95 A0A1B0B834 W8AMF9 A0A0A1X2R8 A0A1I8MV17 Q17P54 A0A1W4WKV8 A0A1I8PK55 A0A1J1J0K1 A0A1B0GHU3 A0A182LCJ9 A0A1B0GIU2 A0A1J1J3M1 Q7QBC5 A0A1I8PK30 A0A182USC5 A0A2C9H8C1 A0A1A9UMF1 A0A336N405 A0A1W4VVW5 A0A453YJK6 N6T454 A0A182IP77 U4UDP3 Q16M91 B4KDU1 A0A2H8TWC3 A0A1A9X1V2 A0A3B0K1B9 B4G4W2 Q298Y7 A0A182L8U0 A0A0L7RHX4 B4PQM8 B4M062 B3P5N5 B4N9K3 B3LXP5 Q9VAR6 B4QYV4 B4HZ33 B4JTC2 A0A437BJF4 Q5TQN9 A0A182NQK4 A0A182Q388 A0A0P8Y1B6 Q17DU8 A0A2S2P3R2 A0A182TD00 A0A182KEE5 A0A182YBE1 E0V9A6 A0A182HKI9 A0A182RNJ6 A0A182M6I9 A0A182X9K6 A0A453Z0K5 A0A026X2K4 J9JYG8 A0A2S2PC71 A0A182VDP8 Q7PXA4 A0A0L0CNQ0 A0A1Y1L5Q2 E1ZV74 A0A084VXU9 A0A182JK23 D6W7P0 A0A2H8TU39 A0A437BJU2 A0A182VVT5 A0A158N9S9 A0A2A4JLN5 A0A158N9A5 A0A194PPY2 A0A1E1W0X2 T1IV88 A0A195CCJ1
A0A1B6DBF6 A0A1B6KXD0 A0A139WNB8 E0VYZ0 D6WCY9 A0A139WN83 A0A1W4XLM5 A0A2A4K566 A0A437B3S3 A0A194QC96 A0A194RDC2 A0A1B0GE93 A0A067RKA0 A0A1A9Z6V7 A0A1A9XQ95 A0A1B0B834 W8AMF9 A0A0A1X2R8 A0A1I8MV17 Q17P54 A0A1W4WKV8 A0A1I8PK55 A0A1J1J0K1 A0A1B0GHU3 A0A182LCJ9 A0A1B0GIU2 A0A1J1J3M1 Q7QBC5 A0A1I8PK30 A0A182USC5 A0A2C9H8C1 A0A1A9UMF1 A0A336N405 A0A1W4VVW5 A0A453YJK6 N6T454 A0A182IP77 U4UDP3 Q16M91 B4KDU1 A0A2H8TWC3 A0A1A9X1V2 A0A3B0K1B9 B4G4W2 Q298Y7 A0A182L8U0 A0A0L7RHX4 B4PQM8 B4M062 B3P5N5 B4N9K3 B3LXP5 Q9VAR6 B4QYV4 B4HZ33 B4JTC2 A0A437BJF4 Q5TQN9 A0A182NQK4 A0A182Q388 A0A0P8Y1B6 Q17DU8 A0A2S2P3R2 A0A182TD00 A0A182KEE5 A0A182YBE1 E0V9A6 A0A182HKI9 A0A182RNJ6 A0A182M6I9 A0A182X9K6 A0A453Z0K5 A0A026X2K4 J9JYG8 A0A2S2PC71 A0A182VDP8 Q7PXA4 A0A0L0CNQ0 A0A1Y1L5Q2 E1ZV74 A0A084VXU9 A0A182JK23 D6W7P0 A0A2H8TU39 A0A437BJU2 A0A182VVT5 A0A158N9S9 A0A2A4JLN5 A0A158N9A5 A0A194PPY2 A0A1E1W0X2 T1IV88 A0A195CCJ1
Pubmed
22118469
26354079
18362917
19820115
20566863
24845553
+ More
24495485 25830018 25315136 17510324 20966253 12364791 14747013 17210077 23537049 17994087 18057021 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30688651 25244985 24508170 26108605 28004739 20798317 24438588 21347285
24495485 25830018 25315136 17510324 20966253 12364791 14747013 17210077 23537049 17994087 18057021 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30688651 25244985 24508170 26108605 28004739 20798317 24438588 21347285
EMBL
RSAL01000128
RVE46511.1
AGBW02009782
OWR49926.1
NWSH01006973
PCG63170.1
+ More
KQ459585 KPI98404.1 KQ460622 KPJ13528.1 KQ971311 EEZ99076.2 GEDC01014275 GEDC01008700 JAS23023.1 JAS28598.1 GEBQ01023864 JAT16113.1 KYB29490.1 DS235848 EEB18596.1 EEZ99078.1 KYB29489.1 NWSH01000118 PCG79381.1 RSAL01000186 RVE44805.1 KQ459193 KPJ03087.1 KQ460325 KPJ15838.1 CCAG010012177 CCAG010012178 KK852460 KDR23453.1 JXJN01009819 GAMC01020882 GAMC01020879 JAB85673.1 GBXI01009247 JAD05045.1 CH477194 EAT48443.1 CVRI01000064 CRL05482.1 AJWK01005625 AJWK01005626 AJWK01005627 AJWK01005628 AJWK01005629 AJWK01005630 AJWK01005631 AJWK01005632 AJWK01005633 AJWK01005634 AJWK01017657 CRL05481.1 AAAB01008879 EAA08340.4 UFQT01002324 SSX33218.1 APCN01002137 APCN01002138 APCN01002139 APCN01002140 APCN01002141 APCN01002142 APGK01044470 APGK01044471 APGK01044472 APGK01044473 APGK01044474 KB741026 ENN74934.1 KB632309 ERL92094.1 CH477872 EAT35448.1 CH933806 EDW14938.2 KRG01288.1 KRG01289.1 GFXV01006037 MBW17842.1 OUUW01000013 SPP88067.1 CH479179 EDW24628.1 CM000070 EAL27817.2 KQ414589 KOC70419.1 CM000160 EDW98367.1 CH940650 EDW68312.2 CH954182 EDV53285.1 CH964232 EDW81679.1 CH902617 EDV43939.1 AE014297 AY060653 AAF56835.1 AAL28201.1 CM000364 EDX14773.1 CH480819 EDW53290.1 CH916373 EDV95012.1 RSAL01000047 RVE50523.1 AAAB01008963 EAL39761.3 AXCN02001517 KPU80550.1 CH477291 EAT44564.1 GGMR01011502 MBY24121.1 DS234991 EEB09962.1 APCN01000831 AXCM01001271 AAAB01008987 KK107024 EZA62236.1 ABLF02039711 ABLF02039721 ABLF02039724 ABLF02039727 ABLF02039728 ABLF02039730 ABLF02047045 GGMR01014375 MBY26994.1 EAA01197.5 JRES01000236 KNC33079.1 GEZM01068577 JAV66886.1 GL434441 EFN74933.1 ATLV01018207 KE525224 KFB42793.1 KQ971307 EFA11252.2 GFXV01005968 MBW17773.1 RVE50525.1 ADTU01009347 ADTU01009348 ADTU01009349 ADTU01009350 ADTU01009351 ADTU01009352 ADTU01009353 ADTU01009354 ADTU01009355 ADTU01009356 NWSH01001156 PCG72322.1 ADTU01009130 ADTU01009131 ADTU01009132 ADTU01009133 ADTU01009134 ADTU01009135 ADTU01009136 ADTU01009137 ADTU01009138 KQ459597 KPI94794.1 GDQN01010475 JAT80579.1 JH431578 KQ977935 KYM98574.1
KQ459585 KPI98404.1 KQ460622 KPJ13528.1 KQ971311 EEZ99076.2 GEDC01014275 GEDC01008700 JAS23023.1 JAS28598.1 GEBQ01023864 JAT16113.1 KYB29490.1 DS235848 EEB18596.1 EEZ99078.1 KYB29489.1 NWSH01000118 PCG79381.1 RSAL01000186 RVE44805.1 KQ459193 KPJ03087.1 KQ460325 KPJ15838.1 CCAG010012177 CCAG010012178 KK852460 KDR23453.1 JXJN01009819 GAMC01020882 GAMC01020879 JAB85673.1 GBXI01009247 JAD05045.1 CH477194 EAT48443.1 CVRI01000064 CRL05482.1 AJWK01005625 AJWK01005626 AJWK01005627 AJWK01005628 AJWK01005629 AJWK01005630 AJWK01005631 AJWK01005632 AJWK01005633 AJWK01005634 AJWK01017657 CRL05481.1 AAAB01008879 EAA08340.4 UFQT01002324 SSX33218.1 APCN01002137 APCN01002138 APCN01002139 APCN01002140 APCN01002141 APCN01002142 APGK01044470 APGK01044471 APGK01044472 APGK01044473 APGK01044474 KB741026 ENN74934.1 KB632309 ERL92094.1 CH477872 EAT35448.1 CH933806 EDW14938.2 KRG01288.1 KRG01289.1 GFXV01006037 MBW17842.1 OUUW01000013 SPP88067.1 CH479179 EDW24628.1 CM000070 EAL27817.2 KQ414589 KOC70419.1 CM000160 EDW98367.1 CH940650 EDW68312.2 CH954182 EDV53285.1 CH964232 EDW81679.1 CH902617 EDV43939.1 AE014297 AY060653 AAF56835.1 AAL28201.1 CM000364 EDX14773.1 CH480819 EDW53290.1 CH916373 EDV95012.1 RSAL01000047 RVE50523.1 AAAB01008963 EAL39761.3 AXCN02001517 KPU80550.1 CH477291 EAT44564.1 GGMR01011502 MBY24121.1 DS234991 EEB09962.1 APCN01000831 AXCM01001271 AAAB01008987 KK107024 EZA62236.1 ABLF02039711 ABLF02039721 ABLF02039724 ABLF02039727 ABLF02039728 ABLF02039730 ABLF02047045 GGMR01014375 MBY26994.1 EAA01197.5 JRES01000236 KNC33079.1 GEZM01068577 JAV66886.1 GL434441 EFN74933.1 ATLV01018207 KE525224 KFB42793.1 KQ971307 EFA11252.2 GFXV01005968 MBW17773.1 RVE50525.1 ADTU01009347 ADTU01009348 ADTU01009349 ADTU01009350 ADTU01009351 ADTU01009352 ADTU01009353 ADTU01009354 ADTU01009355 ADTU01009356 NWSH01001156 PCG72322.1 ADTU01009130 ADTU01009131 ADTU01009132 ADTU01009133 ADTU01009134 ADTU01009135 ADTU01009136 ADTU01009137 ADTU01009138 KQ459597 KPI94794.1 GDQN01010475 JAT80579.1 JH431578 KQ977935 KYM98574.1
Proteomes
UP000283053
UP000007151
UP000218220
UP000053268
UP000053240
UP000007266
+ More
UP000009046 UP000192223 UP000092444 UP000027135 UP000092445 UP000092443 UP000092460 UP000095301 UP000008820 UP000095300 UP000183832 UP000092461 UP000075882 UP000007062 UP000075903 UP000076407 UP000078200 UP000192221 UP000075840 UP000019118 UP000075880 UP000030742 UP000009192 UP000091820 UP000268350 UP000008744 UP000001819 UP000053825 UP000002282 UP000008792 UP000008711 UP000007798 UP000007801 UP000000803 UP000000304 UP000001292 UP000001070 UP000075884 UP000075886 UP000075902 UP000075881 UP000076408 UP000075900 UP000075883 UP000053097 UP000007819 UP000037069 UP000000311 UP000030765 UP000075920 UP000005205 UP000078542
UP000009046 UP000192223 UP000092444 UP000027135 UP000092445 UP000092443 UP000092460 UP000095301 UP000008820 UP000095300 UP000183832 UP000092461 UP000075882 UP000007062 UP000075903 UP000076407 UP000078200 UP000192221 UP000075840 UP000019118 UP000075880 UP000030742 UP000009192 UP000091820 UP000268350 UP000008744 UP000001819 UP000053825 UP000002282 UP000008792 UP000008711 UP000007798 UP000007801 UP000000803 UP000000304 UP000001292 UP000001070 UP000075884 UP000075886 UP000075902 UP000075881 UP000076408 UP000075900 UP000075883 UP000053097 UP000007819 UP000037069 UP000000311 UP000030765 UP000075920 UP000005205 UP000078542
PRIDE
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A3S2NFW5
A0A212F862
A0A2A4IVJ7
A0A194Q082
A0A0N0PCD5
D6WCY6
+ More
A0A1B6DBF6 A0A1B6KXD0 A0A139WNB8 E0VYZ0 D6WCY9 A0A139WN83 A0A1W4XLM5 A0A2A4K566 A0A437B3S3 A0A194QC96 A0A194RDC2 A0A1B0GE93 A0A067RKA0 A0A1A9Z6V7 A0A1A9XQ95 A0A1B0B834 W8AMF9 A0A0A1X2R8 A0A1I8MV17 Q17P54 A0A1W4WKV8 A0A1I8PK55 A0A1J1J0K1 A0A1B0GHU3 A0A182LCJ9 A0A1B0GIU2 A0A1J1J3M1 Q7QBC5 A0A1I8PK30 A0A182USC5 A0A2C9H8C1 A0A1A9UMF1 A0A336N405 A0A1W4VVW5 A0A453YJK6 N6T454 A0A182IP77 U4UDP3 Q16M91 B4KDU1 A0A2H8TWC3 A0A1A9X1V2 A0A3B0K1B9 B4G4W2 Q298Y7 A0A182L8U0 A0A0L7RHX4 B4PQM8 B4M062 B3P5N5 B4N9K3 B3LXP5 Q9VAR6 B4QYV4 B4HZ33 B4JTC2 A0A437BJF4 Q5TQN9 A0A182NQK4 A0A182Q388 A0A0P8Y1B6 Q17DU8 A0A2S2P3R2 A0A182TD00 A0A182KEE5 A0A182YBE1 E0V9A6 A0A182HKI9 A0A182RNJ6 A0A182M6I9 A0A182X9K6 A0A453Z0K5 A0A026X2K4 J9JYG8 A0A2S2PC71 A0A182VDP8 Q7PXA4 A0A0L0CNQ0 A0A1Y1L5Q2 E1ZV74 A0A084VXU9 A0A182JK23 D6W7P0 A0A2H8TU39 A0A437BJU2 A0A182VVT5 A0A158N9S9 A0A2A4JLN5 A0A158N9A5 A0A194PPY2 A0A1E1W0X2 T1IV88 A0A195CCJ1
A0A1B6DBF6 A0A1B6KXD0 A0A139WNB8 E0VYZ0 D6WCY9 A0A139WN83 A0A1W4XLM5 A0A2A4K566 A0A437B3S3 A0A194QC96 A0A194RDC2 A0A1B0GE93 A0A067RKA0 A0A1A9Z6V7 A0A1A9XQ95 A0A1B0B834 W8AMF9 A0A0A1X2R8 A0A1I8MV17 Q17P54 A0A1W4WKV8 A0A1I8PK55 A0A1J1J0K1 A0A1B0GHU3 A0A182LCJ9 A0A1B0GIU2 A0A1J1J3M1 Q7QBC5 A0A1I8PK30 A0A182USC5 A0A2C9H8C1 A0A1A9UMF1 A0A336N405 A0A1W4VVW5 A0A453YJK6 N6T454 A0A182IP77 U4UDP3 Q16M91 B4KDU1 A0A2H8TWC3 A0A1A9X1V2 A0A3B0K1B9 B4G4W2 Q298Y7 A0A182L8U0 A0A0L7RHX4 B4PQM8 B4M062 B3P5N5 B4N9K3 B3LXP5 Q9VAR6 B4QYV4 B4HZ33 B4JTC2 A0A437BJF4 Q5TQN9 A0A182NQK4 A0A182Q388 A0A0P8Y1B6 Q17DU8 A0A2S2P3R2 A0A182TD00 A0A182KEE5 A0A182YBE1 E0V9A6 A0A182HKI9 A0A182RNJ6 A0A182M6I9 A0A182X9K6 A0A453Z0K5 A0A026X2K4 J9JYG8 A0A2S2PC71 A0A182VDP8 Q7PXA4 A0A0L0CNQ0 A0A1Y1L5Q2 E1ZV74 A0A084VXU9 A0A182JK23 D6W7P0 A0A2H8TU39 A0A437BJU2 A0A182VVT5 A0A158N9S9 A0A2A4JLN5 A0A158N9A5 A0A194PPY2 A0A1E1W0X2 T1IV88 A0A195CCJ1
PDB
6EFY
E-value=1.65443e-80,
Score=761
Ontologies
GO
Topology
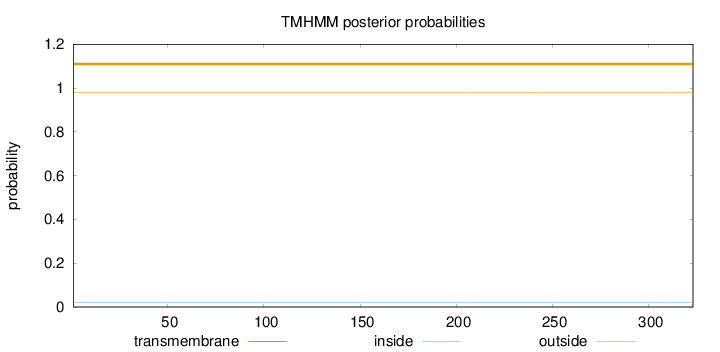
Length:
323
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00645
Exp number, first 60 AAs:
3e-05
Total prob of N-in:
0.02074
outside
1 - 323
Population Genetic Test Statistics
Pi
26.432601
Theta
172.941158
Tajima's D
1.122064
CLR
0.281608
CSRT
0.692715364231788
Interpretation
Uncertain
