Gene
KWMTBOMO12501
Pre Gene Modal
BGIBMGA001503
Annotation
PREDICTED:_uncharacterized_protein_LOC105842132_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.735
Sequence
CDS
ATGGAGGCGGCGTGTCTCAAACGTATGGAGGATCTCAACCAAACCCATCGGAGAACCAGCAATAGTAGAATCCTTACCAAAAAACTCTGCGCCAGCAACAAACAATCTAGCACCAGAATAGGAATCTTACAAATTGAGAACATTTCATACCAGATTACAAACGACTACTCACAAATTGAAACGTTCTACCCATCTCTCAACTTTCTCTACGCCAATGTACGCAGCATTGTTAAGCCGGGAAAGTTTGATGAGCTCAAGTGCGTCCTGAAGTCTATCGATAAGGGTGTACACGTTGTTCTTCTCACCGAAACTTGGATCAAATCTGAAAATGATGCCATGAAACTACATTTGCCAAATTACACTCATCATTACAGCTATCGTAACGACATCAGAGGAGGCGGCGTGTCTATATACGTCCATAATAATATCAAACACAGCACGACTGAAGATAAATACAGCAATGGCAATAACTATCTTTGGGTGTATCTTGAGCAACATGGATTACACGTTGGAGTTGTATACAAGCCAGGAACCACAAATACAGAAGATTTTCTTATCGATTATGAACAGCAACTACATGACAGAAATCGAACCATTATTTTCGGGGATTTCAACCTAGACCTATTACAAGAAGACAAACAAACTAAAATGTATAAACAAATGCTTTGCGAAAATGGATATAAAATATTGAATAAAGTAGAAGAAGAATACTGTACAAGAGAAACGTCGACAACTAGGACAATTTTGGATCATGTTACAACAAATTTAAAGAATAAAAACGTACACTTAGCCATTGTTGAATCTTGTATGTCGGATCATAAACAAATTTACGTCGCAATAAAAAAACAACAAGTAATTTCAAAACAAAGGAGTAAATACGAGGCCATAGACTATAGCAGCCTATATAAAAAATTTGATTCAAGCAAATTAGACAATAGTAACTATGAATACAAACATTTAGAAGATGTTATAAAACAGAATATTCGTGAAAACACTAAGACAAAATCGAAAATACTTAATTTGCCACAACAGGACTGGATAACAAAAAATATCACTGATGGAATAAATAGAAGAAATAAACTGTGGTACAAGCTGAAAAAGGACCCGAATAATGAAGACCTCAAAGCTAATTTCAAAACTGAAAGAAACCAGGTAACGCGGGATATACAAAGCGCAAAGAGAAAATATTATCTGGATGCTTTCAATAAATGTTCCAAAAAACCGAAAAAGATGTGGAGCCTAATCAACACCTTAGCAACAAATAAACTGAAGAAAAAATGTCTGCTTCCCAAGATCCATTCTACTGAGGGTCGTATAATACAAGAAGATCACATATGTGAATATTTTAACAAGTTTTTCTCAACTGTAGGCACAACCTTGGCCGATAAAATTCCTCAGAAATTTCATGGAAATCATACGCATATCTTACCAAATAACTCTGTGACTAATGTTGTTCTACAAAAGCTTAATTTATGTACCTCAGATGAAATTGATAAACATATTGATAACTTGGACCCGAACACAAGTACGGGGATTGATGGTATTAGTGCAAAGACCCTCAAATGTTTAAAGAACGCTGTATGTCTCGTGTTATCACAATGCATTAACAAGTGTTTGAACGATGGTATATTTCCTGATAATCTTAAGATCGCCAAAGTCGTTCCAATTTTCAAAACAGGGTCTAGATCTGATGTTAACAATTATCGTCCCATATCAGTTCTACCCATACCATCTAAAATTTTCGAAAAAGTTCTTTATAATCGTCTCAATGAACACTTAACATCACTGAGTTTTCTTTCTGATCGCCAATACGGGTTTAGGCCAAAATCAAATACACTTTCAGCAACGATAGATCTAGTTACTACTATCAAAACAAACATTGATAAGAAAAACATGGTCTTAGGAGTGTTTATAGACCTAAAAAAGGCTTTTGACACAGTCAGTCACCCATTACTGTTGAAAAAACTAGAGAGCATTGGTGTGACCAATTCAGCTTACGAAATATTTAAGTCTTACTTGGAAAACAGGGTCCAGATGGTGAAAATTGATGATCATCAAAGATCTTTGCGCGGGACCGCCCAGTGTTTTCCTCGTCGAGTCTCTCTCATGATCTACAATTCGTTGGTTAAACCTCATCTTCAATATCTGATTGAGACAGTTACATCCATAGAAATGATAGTATTGTTACTTAAGCCTACGTTAATAACTAAAAATACTAAATTCAATTTAAAGTAA
Protein
MEAACLKRMEDLNQTHRRTSNSRILTKKLCASNKQSSTRIGILQIENISYQITNDYSQIETFYPSLNFLYANVRSIVKPGKFDELKCVLKSIDKGVHVVLLTETWIKSENDAMKLHLPNYTHHYSYRNDIRGGGVSIYVHNNIKHSTTEDKYSNGNNYLWVYLEQHGLHVGVVYKPGTTNTEDFLIDYEQQLHDRNRTIIFGDFNLDLLQEDKQTKMYKQMLCENGYKILNKVEEEYCTRETSTTRTILDHVTTNLKNKNVHLAIVESCMSDHKQIYVAIKKQQVISKQRSKYEAIDYSSLYKKFDSSKLDNSNYEYKHLEDVIKQNIRENTKTKSKILNLPQQDWITKNITDGINRRNKLWYKLKKDPNNEDLKANFKTERNQVTRDIQSAKRKYYLDAFNKCSKKPKKMWSLINTLATNKLKKKCLLPKIHSTEGRIIQEDHICEYFNKFFSTVGTTLADKIPQKFHGNHTHILPNNSVTNVVLQKLNLCTSDEIDKHIDNLDPNTSTGIDGISAKTLKCLKNAVCLVLSQCINKCLNDGIFPDNLKIAKVVPIFKTGSRSDVNNYRPISVLPIPSKIFEKVLYNRLNEHLTSLSFLSDRQYGFRPKSNTLSATIDLVTTIKTNIDKKNMVLGVFIDLKKAFDTVSHPLLLKKLESIGVTNSAYEIFKSYLENRVQMVKIDDHQRSLRGTAQCFPRRVSLMIYNSLVKPHLQYLIETVTSIEMIVLLLKPTLITKNTKFNLK
Summary
Uniprot
H9IW72
A0A2H1VEI6
U5EHK5
L7MML1
L7MA61
L7MAV1
+ More
A0A2A4JXL1 A0A293N5E5 L7MGP0 W4Z536 A0A432I5G5 U5EQD5 A0A3Q3BKX6 A0A131Y4J7 A0A1A8IUC7 A0A146Q9A7 A0A1W7R632 A0A2A4JYZ1 A0A3C1S1K0 U5EEF0 A0A1A8BMQ9 A0A147BM23 L7MAD0 A0A2S2NMQ6 A0A3B3C2G3 A0A1A8MQ07 A0A3B3BEL9 A0A432I6H2 A0A147BN59 A0A3P9L5D0 A0A3P9M519 A0A3B3I8P5 A0A2W1BQJ1 A0A3B5QJ22 A0A147BMB8 A0A1W7R639 A0A0A9WFJ5 L7MDE9 A0A147BK38 A0A293M0N9 A0A2B4SVL4 L7LW06 A0A3B5QM44 A0A432PY41 A0A147BJU8 A0A3B5QVU5 A0A1A8P8T9 A0A2B4RRX2 A0A3P8Q4R8 A0A354GI06 A0A3C1RZ19 A0A147BLD7 A0A2B4SVM1 A0A354GG67 J9KJT6 A0A2G8JVN9 A0A2S2NHB6 A0A2B4SX38 J9JVB9 A0A3P8NF23 A0A3P8N9X1 A0A3B3I7Q5 A0A1E1XIZ4 L7LST3 A0A3Q3JA85 A0A131Y5S4 A0A3B3HKX5 A0A0N7ZAE4 A0A3Q3FLM1 A0A3P8P2B9 A0A293LDA4 A0A1A8GC74 A0A146L3X4
A0A2A4JXL1 A0A293N5E5 L7MGP0 W4Z536 A0A432I5G5 U5EQD5 A0A3Q3BKX6 A0A131Y4J7 A0A1A8IUC7 A0A146Q9A7 A0A1W7R632 A0A2A4JYZ1 A0A3C1S1K0 U5EEF0 A0A1A8BMQ9 A0A147BM23 L7MAD0 A0A2S2NMQ6 A0A3B3C2G3 A0A1A8MQ07 A0A3B3BEL9 A0A432I6H2 A0A147BN59 A0A3P9L5D0 A0A3P9M519 A0A3B3I8P5 A0A2W1BQJ1 A0A3B5QJ22 A0A147BMB8 A0A1W7R639 A0A0A9WFJ5 L7MDE9 A0A147BK38 A0A293M0N9 A0A2B4SVL4 L7LW06 A0A3B5QM44 A0A432PY41 A0A147BJU8 A0A3B5QVU5 A0A1A8P8T9 A0A2B4RRX2 A0A3P8Q4R8 A0A354GI06 A0A3C1RZ19 A0A147BLD7 A0A2B4SVM1 A0A354GG67 J9KJT6 A0A2G8JVN9 A0A2S2NHB6 A0A2B4SX38 J9JVB9 A0A3P8NF23 A0A3P8N9X1 A0A3B3I7Q5 A0A1E1XIZ4 L7LST3 A0A3Q3JA85 A0A131Y5S4 A0A3B3HKX5 A0A0N7ZAE4 A0A3Q3FLM1 A0A3P8P2B9 A0A293LDA4 A0A1A8GC74 A0A146L3X4
Pubmed
EMBL
BABH01005955
BABH01005956
BABH01005957
ODYU01002150
SOQ39280.1
GANO01002979
+ More
JAB56892.1 GACK01000380 JAA64654.1 GACK01003878 JAA61156.1 GACK01003889 JAA61145.1 NWSH01000417 PCG76539.1 GFWV01022122 MAA46850.1 GACK01002596 JAA62438.1 AAGJ04008561 QNYJ01000098 RUA05525.1 GANO01004336 JAB55535.1 GEFM01002391 JAP73405.1 HAED01014535 SBR00980.1 GCES01133440 JAQ52882.1 GEHC01001019 JAV46626.1 NWSH01000358 PCG77029.1 DMQF01000155 HAO14484.1 GANO01004337 JAB55534.1 HADZ01003859 SBP67800.1 GEGO01003580 JAR91824.1 GACK01003803 JAA61231.1 GGMR01005613 MBY18232.1 HAEF01017628 SBR58787.1 QNYJ01000080 RUA05885.1 GEGO01003176 JAR92228.1 KZ149952 PZC76581.1 GEGO01003474 JAR91930.1 GEHC01001016 JAV46629.1 GBHO01036382 JAG07222.1 GACK01003815 JAA61219.1 GEGO01004245 JAR91159.1 GFWV01009040 GFWV01009041 MAA33769.1 LSMT01000021 PFX32582.1 GACK01009044 JAA55990.1 QNXQ01000100 RUM30450.1 GEGO01004368 JAR91036.1 HAEG01006811 SBR77686.1 LSMT01000382 PFX18982.1 DNUT01000355 HBI40644.1 DMQF01000079 HAO14226.1 GEGO01003806 JAR91598.1 LSMT01000002 PFX34724.1 DNUT01000184 HBI40005.1 ABLF02015076 ABLF02020029 ABLF02067202 MRZV01001199 PIK39803.1 GGMR01003557 MBY16176.1 LSMT01000016 PFX32997.1 ABLF02011184 ABLF02011185 GFAC01000112 JAT99076.1 GACK01010154 JAA54880.1 GEFM01002390 JAP73406.1 GDRN01100198 JAI58616.1 GFWV01001118 MAA25848.1 HAEC01001258 SBQ69335.1 GDHC01015476 JAQ03153.1
JAB56892.1 GACK01000380 JAA64654.1 GACK01003878 JAA61156.1 GACK01003889 JAA61145.1 NWSH01000417 PCG76539.1 GFWV01022122 MAA46850.1 GACK01002596 JAA62438.1 AAGJ04008561 QNYJ01000098 RUA05525.1 GANO01004336 JAB55535.1 GEFM01002391 JAP73405.1 HAED01014535 SBR00980.1 GCES01133440 JAQ52882.1 GEHC01001019 JAV46626.1 NWSH01000358 PCG77029.1 DMQF01000155 HAO14484.1 GANO01004337 JAB55534.1 HADZ01003859 SBP67800.1 GEGO01003580 JAR91824.1 GACK01003803 JAA61231.1 GGMR01005613 MBY18232.1 HAEF01017628 SBR58787.1 QNYJ01000080 RUA05885.1 GEGO01003176 JAR92228.1 KZ149952 PZC76581.1 GEGO01003474 JAR91930.1 GEHC01001016 JAV46629.1 GBHO01036382 JAG07222.1 GACK01003815 JAA61219.1 GEGO01004245 JAR91159.1 GFWV01009040 GFWV01009041 MAA33769.1 LSMT01000021 PFX32582.1 GACK01009044 JAA55990.1 QNXQ01000100 RUM30450.1 GEGO01004368 JAR91036.1 HAEG01006811 SBR77686.1 LSMT01000382 PFX18982.1 DNUT01000355 HBI40644.1 DMQF01000079 HAO14226.1 GEGO01003806 JAR91598.1 LSMT01000002 PFX34724.1 DNUT01000184 HBI40005.1 ABLF02015076 ABLF02020029 ABLF02067202 MRZV01001199 PIK39803.1 GGMR01003557 MBY16176.1 LSMT01000016 PFX32997.1 ABLF02011184 ABLF02011185 GFAC01000112 JAT99076.1 GACK01010154 JAA54880.1 GEFM01002390 JAP73406.1 GDRN01100198 JAI58616.1 GFWV01001118 MAA25848.1 HAEC01001258 SBQ69335.1 GDHC01015476 JAQ03153.1
Proteomes
Pfam
Interpro
IPR005135
Endo/exonuclease/phosphatase
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR000048 IQ_motif_EF-hand-BS
IPR021109 Peptidase_aspartic_dom_sf
IPR040676 DUF5641
IPR041373 RT_RNaseH
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR011011 Znf_FYVE_PHD
IPR019786 Zinc_finger_PHD-type_CS
IPR018289 MULE_transposase_dom
IPR004941 Baculo_FP
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR019787 Znf_PHD-finger
IPR000859 CUB_dom
IPR035914 Sperma_CUB_dom_sf
IPR007527 Znf_SWIM
IPR011604 Exonuc_phg/RecB_C
IPR011335 Restrct_endonuc-II-like
IPR019080 YqaJ_viral_recombinase
IPR029526 PGBD
IPR008906 HATC_C_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR000048 IQ_motif_EF-hand-BS
IPR021109 Peptidase_aspartic_dom_sf
IPR040676 DUF5641
IPR041373 RT_RNaseH
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR011011 Znf_FYVE_PHD
IPR019786 Zinc_finger_PHD-type_CS
IPR018289 MULE_transposase_dom
IPR004941 Baculo_FP
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR019787 Znf_PHD-finger
IPR000859 CUB_dom
IPR035914 Sperma_CUB_dom_sf
IPR007527 Znf_SWIM
IPR011604 Exonuc_phg/RecB_C
IPR011335 Restrct_endonuc-II-like
IPR019080 YqaJ_viral_recombinase
IPR029526 PGBD
IPR008906 HATC_C_dom
SUPFAM
CDD
ProteinModelPortal
H9IW72
A0A2H1VEI6
U5EHK5
L7MML1
L7MA61
L7MAV1
+ More
A0A2A4JXL1 A0A293N5E5 L7MGP0 W4Z536 A0A432I5G5 U5EQD5 A0A3Q3BKX6 A0A131Y4J7 A0A1A8IUC7 A0A146Q9A7 A0A1W7R632 A0A2A4JYZ1 A0A3C1S1K0 U5EEF0 A0A1A8BMQ9 A0A147BM23 L7MAD0 A0A2S2NMQ6 A0A3B3C2G3 A0A1A8MQ07 A0A3B3BEL9 A0A432I6H2 A0A147BN59 A0A3P9L5D0 A0A3P9M519 A0A3B3I8P5 A0A2W1BQJ1 A0A3B5QJ22 A0A147BMB8 A0A1W7R639 A0A0A9WFJ5 L7MDE9 A0A147BK38 A0A293M0N9 A0A2B4SVL4 L7LW06 A0A3B5QM44 A0A432PY41 A0A147BJU8 A0A3B5QVU5 A0A1A8P8T9 A0A2B4RRX2 A0A3P8Q4R8 A0A354GI06 A0A3C1RZ19 A0A147BLD7 A0A2B4SVM1 A0A354GG67 J9KJT6 A0A2G8JVN9 A0A2S2NHB6 A0A2B4SX38 J9JVB9 A0A3P8NF23 A0A3P8N9X1 A0A3B3I7Q5 A0A1E1XIZ4 L7LST3 A0A3Q3JA85 A0A131Y5S4 A0A3B3HKX5 A0A0N7ZAE4 A0A3Q3FLM1 A0A3P8P2B9 A0A293LDA4 A0A1A8GC74 A0A146L3X4
A0A2A4JXL1 A0A293N5E5 L7MGP0 W4Z536 A0A432I5G5 U5EQD5 A0A3Q3BKX6 A0A131Y4J7 A0A1A8IUC7 A0A146Q9A7 A0A1W7R632 A0A2A4JYZ1 A0A3C1S1K0 U5EEF0 A0A1A8BMQ9 A0A147BM23 L7MAD0 A0A2S2NMQ6 A0A3B3C2G3 A0A1A8MQ07 A0A3B3BEL9 A0A432I6H2 A0A147BN59 A0A3P9L5D0 A0A3P9M519 A0A3B3I8P5 A0A2W1BQJ1 A0A3B5QJ22 A0A147BMB8 A0A1W7R639 A0A0A9WFJ5 L7MDE9 A0A147BK38 A0A293M0N9 A0A2B4SVL4 L7LW06 A0A3B5QM44 A0A432PY41 A0A147BJU8 A0A3B5QVU5 A0A1A8P8T9 A0A2B4RRX2 A0A3P8Q4R8 A0A354GI06 A0A3C1RZ19 A0A147BLD7 A0A2B4SVM1 A0A354GG67 J9KJT6 A0A2G8JVN9 A0A2S2NHB6 A0A2B4SX38 J9JVB9 A0A3P8NF23 A0A3P8N9X1 A0A3B3I7Q5 A0A1E1XIZ4 L7LST3 A0A3Q3JA85 A0A131Y5S4 A0A3B3HKX5 A0A0N7ZAE4 A0A3Q3FLM1 A0A3P8P2B9 A0A293LDA4 A0A1A8GC74 A0A146L3X4
Ontologies
GO
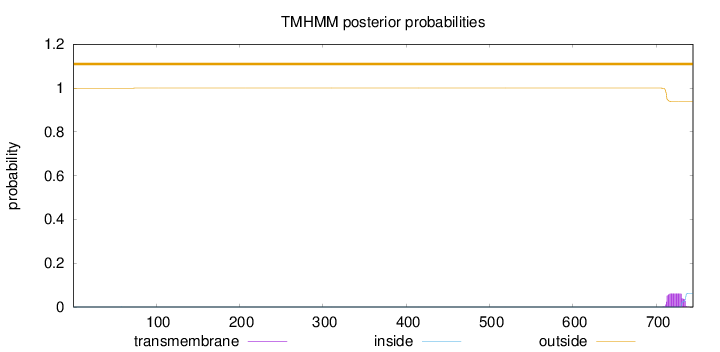
Topology
Length:
744
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.30389
Exp number, first 60 AAs:
0.00033
Total prob of N-in:
0.00039
outside
1 - 744
Population Genetic Test Statistics
Pi
227.736515
Theta
198.0596
Tajima's D
3.092007
CLR
0.159305
CSRT
0.982600869956502
Interpretation
Possibly Balancing Selection
