Pre Gene Modal
BGIBMGA001502
Annotation
geranylgeranyltransferase_type_I_beta_subunit_[Bombyx_mori]
Full name
Geranylgeranyl transferase type-1 subunit beta
Alternative Name
Geranylgeranyl transferase type I subunit beta
Type I protein geranyl-geranyltransferase subunit beta
Type I protein geranyl-geranyltransferase subunit beta
Location in the cell
Extracellular Reliability : 1.538 PlasmaMembrane Reliability : 1.724
Sequence
CDS
ATGAATAACGAAGGAAACAAGGATATGGCTCATCGTCAGCACGTAAAATATTTCATGAGATTTTTGAATATTTTGCCTTCGTCTCTATCATCACACGATACTACAAGAGTTACAATAGCATATTTCTCTGTTGCCGGCCTGGATGTCTTAGGATCCATATCAGCAATATCTCTTGATCTGAGATCCAGGATTATTGAATGGTTGTATAGGCTTCAAGTACATCCTGATAAGGAAACGGGCGACATGTCACTATGCGGCTTCCAAGGCTCCTCAACGATAAACATAAGACTGGATCCGGATAACAATCAATTTCGATGCGGCCATCTAGCGATGACGTACACTGGTCTGTGCATACTGCTGGCGTTGGGCGATGACCTATCGAGAATCAACAGAACAGCCTTAATTCAAGGCGTGAAAGCGTTGCAGACGGACGAAGGGAACTTCTCGGCAACCCTGTCGGGCTGCGAGTCGGACATGCGGTTCGTGTATTGCGCTGCGTGTATCAGTTACATATTGAACGATTGGTCCGGCTTCGATATAGAGAAGGCCACAGATTACGTCATTAAATCTATAGGCTACGACTATGGCATCGCTCAATGTCCCGAGCTAGAATCCCACGGGGGTACGACGTTTTGTGCTCTGGCAACACTAGCGCTAACCGACCAATTGGACAAGCTATCCGAAGCGCAGATCGACGGTCTGAAGCGTTGGTTGGTTTACCGGCAAATCGACGGTTTTCAGGGGAGACCGAATAAGCCGGTCGACACGTGCTACAGTTTCTGGGTGGGCGCCTCGTTGAAGATACTGAACGCTTTACAGCTGACGAACTACGGCAGCAATAGGAGGTACGTGTACGAGACGCAGGATATGGTCGTGGGGGGTTTCTCGAAATGGCCGGACACGTGCACGGACCCCATGCACACGTATTTGGGCCTGTCCGGTTTGAGTTTGATAGGCGAGAGCGGTCTTCTAGAGATAGAACCAAGATTGAACATAACGAAACGGGCCTACGAACATTTGAAGTGTCTGCACGAACAGTGGGTGTCGTAG
Protein
MNNEGNKDMAHRQHVKYFMRFLNILPSSLSSHDTTRVTIAYFSVAGLDVLGSISAISLDLRSRIIEWLYRLQVHPDKETGDMSLCGFQGSSTINIRLDPDNNQFRCGHLAMTYTGLCILLALGDDLSRINRTALIQGVKALQTDEGNFSATLSGCESDMRFVYCAACISYILNDWSGFDIEKATDYVIKSIGYDYGIAQCPELESHGGTTFCALATLALTDQLDKLSEAQIDGLKRWLVYRQIDGFQGRPNKPVDTCYSFWVGASLKILNALQLTNYGSNRRYVYETQDMVVGGFSKWPDTCTDPMHTYLGLSGLSLIGESGLLEIEPRLNITKRAYEHLKCLHEQWVS
Summary
Description
Catalyzes the transfer of a geranyl-geranyl moiety from geranyl-geranyl pyrophosphate to a cysteine at the fourth position from the C-terminus of proteins having the C-terminal sequence Cys-aliphatic-aliphatic-X. Known substrates include RAC1, RAC2, RAP1A and RAP1B (By similarity).
Catalytic Activity
geranylgeranyl diphosphate + L-cysteinyl-[protein] = diphosphate + S-geranylgeranyl-L-cysteinyl-[protein]
Cofactor
Zn(2+)
Subunit
Heterodimer of FNTA and PGGT1B. PGGT1B mediates interaction with substrate peptides (By similarity).
Similarity
Belongs to the protein prenyltransferase subunit beta family.
Keywords
Complete proteome
Metal-binding
Prenyltransferase
Reference proteome
Repeat
Transferase
Zinc
Feature
chain Geranylgeranyl transferase type-1 subunit beta
Uniprot
D6RVZ4
A0A2A4K3P8
A0A2H1VLZ2
A0A0N0PB21
A0A194PZD2
A0A212F8B3
+ More
H9IW71 A0A067RGQ2 A0A0K8TS57 A0A2J7PZS7 A0A084WH47 A0A2M4BRL2 A0A182F3H7 W5JAS0 A0A2M4AR28 A0A2M4CI48 A0A2M4BSG2 A0A2M4BRN7 Q17K57 A0A182M0T3 A0A182VUE6 A0A1Q3FCJ0 A0A1B6LQY0 A0A1Q3FCE5 A0A224XF49 B0XBT8 A0A182IUM4 A0A182YDS6 A0A023EQT2 A0A182RIR4 A0A182K8A7 A0A182L0H4 A0A069DTC3 A0A182SN11 A0A182HEH4 A0A182QX37 A0A182VD82 A0A1Q3FCI3 A0A1B6I2N9 A0A182U3Y5 Q7Q4H9 A0A182IC45 A0A182XFW1 A0A182PLT3 A0A182NU16 A0A1Y1KZU9 R4FLG1 A0A1L8DU25 A0A1B6GZH9 T1E9L3 A0A2R7VW16 A0A0L0C702 A0A0F7Z9D9 T1DBS6 A0A0B8RXM5 A0A2H6N614 A7SXB0 A0A1I8N947 W8BKP7 A0A1I8PKF4 A0A034VHI2 A0A0K8ULX0 A0A2Y9DLS1 A0A0A9WT78 A0A402FDF6 A0A384DL33 G1L1G2 A0A3Q7XAV5 D2HGS9 A0A0K8SRS5 A0A218UU77 H0YSW7 A0A1S3PK25 E2QZU5 A0A3Q7T2L3 A0A0Q9WBJ4 A0A0A1X7I4 G3ST05 B4LS59 A0A060W4I7 A0A3B4CCS5 F7BSD0 L5KLY1 A0A1S3W9P7 A0A369S4S4 A0A0Q3LTJ8 A0A286XNF3 W5MRC0 A0A341BAC8 A0A2U4BT74 A0A2Y9PLG6 Q5EAD5 A0A452G303 F6W1R0 A0A1A9UPJ5 A0A340WR36 A0A2Y9F6M6 M3X221 A0A2Y9I0N3 G1PXD6
H9IW71 A0A067RGQ2 A0A0K8TS57 A0A2J7PZS7 A0A084WH47 A0A2M4BRL2 A0A182F3H7 W5JAS0 A0A2M4AR28 A0A2M4CI48 A0A2M4BSG2 A0A2M4BRN7 Q17K57 A0A182M0T3 A0A182VUE6 A0A1Q3FCJ0 A0A1B6LQY0 A0A1Q3FCE5 A0A224XF49 B0XBT8 A0A182IUM4 A0A182YDS6 A0A023EQT2 A0A182RIR4 A0A182K8A7 A0A182L0H4 A0A069DTC3 A0A182SN11 A0A182HEH4 A0A182QX37 A0A182VD82 A0A1Q3FCI3 A0A1B6I2N9 A0A182U3Y5 Q7Q4H9 A0A182IC45 A0A182XFW1 A0A182PLT3 A0A182NU16 A0A1Y1KZU9 R4FLG1 A0A1L8DU25 A0A1B6GZH9 T1E9L3 A0A2R7VW16 A0A0L0C702 A0A0F7Z9D9 T1DBS6 A0A0B8RXM5 A0A2H6N614 A7SXB0 A0A1I8N947 W8BKP7 A0A1I8PKF4 A0A034VHI2 A0A0K8ULX0 A0A2Y9DLS1 A0A0A9WT78 A0A402FDF6 A0A384DL33 G1L1G2 A0A3Q7XAV5 D2HGS9 A0A0K8SRS5 A0A218UU77 H0YSW7 A0A1S3PK25 E2QZU5 A0A3Q7T2L3 A0A0Q9WBJ4 A0A0A1X7I4 G3ST05 B4LS59 A0A060W4I7 A0A3B4CCS5 F7BSD0 L5KLY1 A0A1S3W9P7 A0A369S4S4 A0A0Q3LTJ8 A0A286XNF3 W5MRC0 A0A341BAC8 A0A2U4BT74 A0A2Y9PLG6 Q5EAD5 A0A452G303 F6W1R0 A0A1A9UPJ5 A0A340WR36 A0A2Y9F6M6 M3X221 A0A2Y9I0N3 G1PXD6
EC Number
2.5.1.59
Pubmed
26354079
22118469
19121390
24845553
26369729
24438588
+ More
20920257 23761445 17510324 25244985 24945155 20966253 26334808 26483478 12364791 14747013 17210077 28004739 26108605 23758969 25727380 25476704 17615350 25315136 24495485 25348373 25401762 26823975 24813606 20010809 20360741 16341006 17994087 25830018 24755649 17495919 23258410 30042472 21993624 16305752 19892987 17975172
20920257 23761445 17510324 25244985 24945155 20966253 26334808 26483478 12364791 14747013 17210077 28004739 26108605 23758969 25727380 25476704 17615350 25315136 24495485 25348373 25401762 26823975 24813606 20010809 20360741 16341006 17994087 25830018 24755649 17495919 23258410 30042472 21993624 16305752 19892987 17975172
EMBL
AB568272
BAJ09607.1
NWSH01000166
PCG78885.1
ODYU01003290
SOQ41855.1
+ More
KQ461068 KPJ09878.1 KQ459585 KPI98403.1 AGBW02009774 OWR49949.1 BABH01005937 KK852693 KDR18308.1 GDAI01000414 JAI17189.1 NEVH01020332 PNF21830.1 ATLV01023789 KE525346 KFB49541.1 GGFJ01006586 MBW55727.1 ADMH02001920 ETN60498.1 GGFK01009925 MBW43246.1 GGFL01000836 MBW65014.1 GGFJ01006587 MBW55728.1 GGFJ01006588 MBW55729.1 CH477228 EAT47035.1 AXCM01003842 GFDL01009750 JAV25295.1 GEBQ01013826 GEBQ01009402 JAT26151.1 JAT30575.1 GFDL01009887 JAV25158.1 GFTR01005330 JAW11096.1 DS232653 EDS44467.1 GAPW01002135 JAC11463.1 GBGD01001992 JAC86897.1 JXUM01131599 JXUM01131600 KQ567733 KXJ69303.1 AXCN02000809 GFDL01009816 JAV25229.1 GECU01026510 JAS81196.1 AAAB01008964 EAA12255.4 APCN01000775 GEZM01068580 JAV66884.1 ACPB03003214 GAHY01002124 JAA75386.1 GFDF01004143 JAV09941.1 GECZ01001936 JAS67833.1 GAMD01001247 JAB00344.1 KK854121 PTY11652.1 JRES01000827 KNC28050.1 GBEX01001354 JAI13206.1 GAAZ01000959 GBKC01001429 GBKD01000865 JAA96984.1 JAG44641.1 JAG46753.1 GBSH01001148 JAG67877.1 IACI01056842 LAA25406.1 DS469882 EDO31665.1 GAMC01007214 GAMC01007213 JAB99342.1 GAKP01017370 GAKP01017368 GAKP01017366 JAC41582.1 GDHF01024622 JAI27692.1 GBHO01032590 GDHC01003939 JAG11014.1 JAQ14690.1 BDOT01000203 GCF54493.1 ACTA01043943 ACTA01051943 GL192821 EFB19790.1 GBRD01009762 JAG56062.1 MUZQ01000127 OWK57295.1 ABQF01045107 AAEX03007707 CH940649 KRF81842.1 GBXI01007190 GBXI01002049 JAD07102.1 JAD12243.1 EDW64745.1 FR904341 CDQ60159.1 KB030673 ELK11573.1 NOWV01000075 RDD41434.1 LMAW01003152 KQK73725.1 AAKN02023039 AAKN02023040 AHAT01002849 BT020634 LWLT01000010 AANG04002524 AAPE02000259
KQ461068 KPJ09878.1 KQ459585 KPI98403.1 AGBW02009774 OWR49949.1 BABH01005937 KK852693 KDR18308.1 GDAI01000414 JAI17189.1 NEVH01020332 PNF21830.1 ATLV01023789 KE525346 KFB49541.1 GGFJ01006586 MBW55727.1 ADMH02001920 ETN60498.1 GGFK01009925 MBW43246.1 GGFL01000836 MBW65014.1 GGFJ01006587 MBW55728.1 GGFJ01006588 MBW55729.1 CH477228 EAT47035.1 AXCM01003842 GFDL01009750 JAV25295.1 GEBQ01013826 GEBQ01009402 JAT26151.1 JAT30575.1 GFDL01009887 JAV25158.1 GFTR01005330 JAW11096.1 DS232653 EDS44467.1 GAPW01002135 JAC11463.1 GBGD01001992 JAC86897.1 JXUM01131599 JXUM01131600 KQ567733 KXJ69303.1 AXCN02000809 GFDL01009816 JAV25229.1 GECU01026510 JAS81196.1 AAAB01008964 EAA12255.4 APCN01000775 GEZM01068580 JAV66884.1 ACPB03003214 GAHY01002124 JAA75386.1 GFDF01004143 JAV09941.1 GECZ01001936 JAS67833.1 GAMD01001247 JAB00344.1 KK854121 PTY11652.1 JRES01000827 KNC28050.1 GBEX01001354 JAI13206.1 GAAZ01000959 GBKC01001429 GBKD01000865 JAA96984.1 JAG44641.1 JAG46753.1 GBSH01001148 JAG67877.1 IACI01056842 LAA25406.1 DS469882 EDO31665.1 GAMC01007214 GAMC01007213 JAB99342.1 GAKP01017370 GAKP01017368 GAKP01017366 JAC41582.1 GDHF01024622 JAI27692.1 GBHO01032590 GDHC01003939 JAG11014.1 JAQ14690.1 BDOT01000203 GCF54493.1 ACTA01043943 ACTA01051943 GL192821 EFB19790.1 GBRD01009762 JAG56062.1 MUZQ01000127 OWK57295.1 ABQF01045107 AAEX03007707 CH940649 KRF81842.1 GBXI01007190 GBXI01002049 JAD07102.1 JAD12243.1 EDW64745.1 FR904341 CDQ60159.1 KB030673 ELK11573.1 NOWV01000075 RDD41434.1 LMAW01003152 KQK73725.1 AAKN02023039 AAKN02023040 AHAT01002849 BT020634 LWLT01000010 AANG04002524 AAPE02000259
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000005204
UP000027135
+ More
UP000235965 UP000030765 UP000069272 UP000000673 UP000008820 UP000075883 UP000075920 UP000002320 UP000075880 UP000076408 UP000075900 UP000075881 UP000075882 UP000075901 UP000069940 UP000249989 UP000075886 UP000075903 UP000075902 UP000007062 UP000075840 UP000076407 UP000075885 UP000075884 UP000015103 UP000037069 UP000001593 UP000095301 UP000095300 UP000248480 UP000288954 UP000261680 UP000291021 UP000008912 UP000286642 UP000197619 UP000007754 UP000087266 UP000002254 UP000286640 UP000008792 UP000007646 UP000193380 UP000261440 UP000002280 UP000010552 UP000079721 UP000253843 UP000051836 UP000005447 UP000018468 UP000252040 UP000245320 UP000248483 UP000009136 UP000291000 UP000002281 UP000078200 UP000265300 UP000248484 UP000011712 UP000248481 UP000001074
UP000235965 UP000030765 UP000069272 UP000000673 UP000008820 UP000075883 UP000075920 UP000002320 UP000075880 UP000076408 UP000075900 UP000075881 UP000075882 UP000075901 UP000069940 UP000249989 UP000075886 UP000075903 UP000075902 UP000007062 UP000075840 UP000076407 UP000075885 UP000075884 UP000015103 UP000037069 UP000001593 UP000095301 UP000095300 UP000248480 UP000288954 UP000261680 UP000291021 UP000008912 UP000286642 UP000197619 UP000007754 UP000087266 UP000002254 UP000286640 UP000008792 UP000007646 UP000193380 UP000261440 UP000002280 UP000010552 UP000079721 UP000253843 UP000051836 UP000005447 UP000018468 UP000252040 UP000245320 UP000248483 UP000009136 UP000291000 UP000002281 UP000078200 UP000265300 UP000248484 UP000011712 UP000248481 UP000001074
Pfam
PF00432 Prenyltrans
Interpro
SUPFAM
SSF48239
SSF48239
CDD
ProteinModelPortal
D6RVZ4
A0A2A4K3P8
A0A2H1VLZ2
A0A0N0PB21
A0A194PZD2
A0A212F8B3
+ More
H9IW71 A0A067RGQ2 A0A0K8TS57 A0A2J7PZS7 A0A084WH47 A0A2M4BRL2 A0A182F3H7 W5JAS0 A0A2M4AR28 A0A2M4CI48 A0A2M4BSG2 A0A2M4BRN7 Q17K57 A0A182M0T3 A0A182VUE6 A0A1Q3FCJ0 A0A1B6LQY0 A0A1Q3FCE5 A0A224XF49 B0XBT8 A0A182IUM4 A0A182YDS6 A0A023EQT2 A0A182RIR4 A0A182K8A7 A0A182L0H4 A0A069DTC3 A0A182SN11 A0A182HEH4 A0A182QX37 A0A182VD82 A0A1Q3FCI3 A0A1B6I2N9 A0A182U3Y5 Q7Q4H9 A0A182IC45 A0A182XFW1 A0A182PLT3 A0A182NU16 A0A1Y1KZU9 R4FLG1 A0A1L8DU25 A0A1B6GZH9 T1E9L3 A0A2R7VW16 A0A0L0C702 A0A0F7Z9D9 T1DBS6 A0A0B8RXM5 A0A2H6N614 A7SXB0 A0A1I8N947 W8BKP7 A0A1I8PKF4 A0A034VHI2 A0A0K8ULX0 A0A2Y9DLS1 A0A0A9WT78 A0A402FDF6 A0A384DL33 G1L1G2 A0A3Q7XAV5 D2HGS9 A0A0K8SRS5 A0A218UU77 H0YSW7 A0A1S3PK25 E2QZU5 A0A3Q7T2L3 A0A0Q9WBJ4 A0A0A1X7I4 G3ST05 B4LS59 A0A060W4I7 A0A3B4CCS5 F7BSD0 L5KLY1 A0A1S3W9P7 A0A369S4S4 A0A0Q3LTJ8 A0A286XNF3 W5MRC0 A0A341BAC8 A0A2U4BT74 A0A2Y9PLG6 Q5EAD5 A0A452G303 F6W1R0 A0A1A9UPJ5 A0A340WR36 A0A2Y9F6M6 M3X221 A0A2Y9I0N3 G1PXD6
H9IW71 A0A067RGQ2 A0A0K8TS57 A0A2J7PZS7 A0A084WH47 A0A2M4BRL2 A0A182F3H7 W5JAS0 A0A2M4AR28 A0A2M4CI48 A0A2M4BSG2 A0A2M4BRN7 Q17K57 A0A182M0T3 A0A182VUE6 A0A1Q3FCJ0 A0A1B6LQY0 A0A1Q3FCE5 A0A224XF49 B0XBT8 A0A182IUM4 A0A182YDS6 A0A023EQT2 A0A182RIR4 A0A182K8A7 A0A182L0H4 A0A069DTC3 A0A182SN11 A0A182HEH4 A0A182QX37 A0A182VD82 A0A1Q3FCI3 A0A1B6I2N9 A0A182U3Y5 Q7Q4H9 A0A182IC45 A0A182XFW1 A0A182PLT3 A0A182NU16 A0A1Y1KZU9 R4FLG1 A0A1L8DU25 A0A1B6GZH9 T1E9L3 A0A2R7VW16 A0A0L0C702 A0A0F7Z9D9 T1DBS6 A0A0B8RXM5 A0A2H6N614 A7SXB0 A0A1I8N947 W8BKP7 A0A1I8PKF4 A0A034VHI2 A0A0K8ULX0 A0A2Y9DLS1 A0A0A9WT78 A0A402FDF6 A0A384DL33 G1L1G2 A0A3Q7XAV5 D2HGS9 A0A0K8SRS5 A0A218UU77 H0YSW7 A0A1S3PK25 E2QZU5 A0A3Q7T2L3 A0A0Q9WBJ4 A0A0A1X7I4 G3ST05 B4LS59 A0A060W4I7 A0A3B4CCS5 F7BSD0 L5KLY1 A0A1S3W9P7 A0A369S4S4 A0A0Q3LTJ8 A0A286XNF3 W5MRC0 A0A341BAC8 A0A2U4BT74 A0A2Y9PLG6 Q5EAD5 A0A452G303 F6W1R0 A0A1A9UPJ5 A0A340WR36 A0A2Y9F6M6 M3X221 A0A2Y9I0N3 G1PXD6
PDB
1TNZ
E-value=1.4356e-90,
Score=848
Ontologies
GO
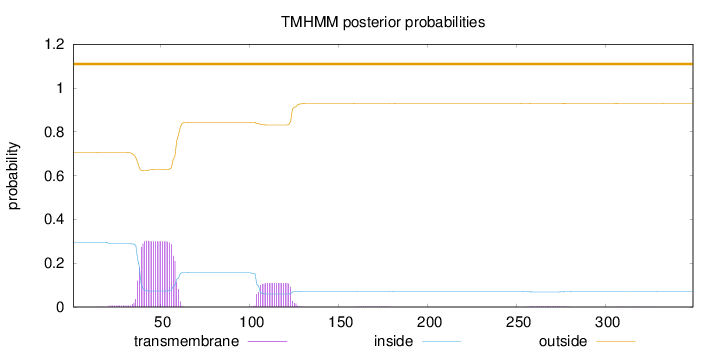
Topology
Length:
349
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.72927000000001
Exp number, first 60 AAs:
6.43845
Total prob of N-in:
0.29488
outside
1 - 349
Population Genetic Test Statistics
Pi
303.427455
Theta
185.423537
Tajima's D
0.707305
CLR
0.310617
CSRT
0.570121493925304
Interpretation
Uncertain
