Gene
KWMTBOMO12480
Pre Gene Modal
BGIBMGA001473
Annotation
PREDICTED:_bumetanide-sensitive_sodium-(potassium)-chloride_cotransporter-like_isoform_X1_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.874
Sequence
CDS
ATGGACGTACGTTTGCTAGGGGACTGTGGGAAAATGGTTCCCAAATCAATGTTGAGACGCAGGCGAAGTGAGTCTGTCGAAATCGGAAACACTAATTGCGAAGTAAATTCCGAAGAAAACGAGAGCAGAAGCACTCGGGAATATAGCAATGGACATTTGCATCCTACTCCGGAACTAACAACGTATGGAGGGATAAAATTAGGATGGATTCAGGGTGTATTGATACCATGTCTCCTCAACATTTGGGGTGTTATGTTGTTTTTACGGATCGCGTGGGTCGTGGCGCAAGCTGGTATAGGTCTTTCGGTCATAATTGTATTCCTTTCCGCCATAGTCTGCGTTATCACGACTCTGTCTCTGAGCGCAATATGCACCAACGGACAACTACAAGGAGGTGGAGTATACTATCTTGTCTCGAGATCACTGGGGGCCGAATTTGGGGCGTCAGTCGGAATACTCTTTGCATTCGCCAACGCTGTAGCGGCCAGTATGAACACTATCGGATTTTGTGATTCTCTAAATCACCTCCTGAAGACGCACGGACTGAAAATCGTTGATAACGACGTAAACGATATCAGGATTGTGGGTGCGGTAGCGCTTTTAGTTATGTGCGTAATTTGTGCCATTGGTATGGACTGGGAAACCAAAGCGCAGAATTTTCTAATAGTCGTAATAGTAGTGGCGATAATAAATTACATTGTCGGAGCACTTATAGGACCCAGTAGTGATGTAGAAAGAGCAAAAGGATTTGTGGGAATAAATTTACAAACTATTCGGGAGAACTGGCTCCCGAATTTTCGCTACAGCGAAGGGCGCGAACACGACTTCTTCAGTGTTTTCGCGGTTTACTTCCCCGCCGTGACTGGCGTGCAGGCGGGGGCCAACATTTGTGGAGATCTCAAGAACCCAGCCGTCGCAATTCCCAAAGGTACTCTTTTAGCGCTGGCCATATCAGTCACGTCGTATTTCACAATGGTGATTCTGTCGGGAATGGGTGCCCTCAGGGACGCGAGCGGTAACGTCGCGAACTTGACAACAGCCGCGCTCGAATCTTGCAAACCTCACTGCGGTTACGGCTTACACAATAACTATGCGATCATGCAGTTAATGTCGGCTTGGGGCCCCTTCATCTACGCGGGCTGCTGGGCGGCGACTCTGTCCACGGCGCTCACCAACTTGCTGTCGGTGCCGCGGCTCATCCAGGCGCTCGGCGTCGACCGCATCTACCCAGGACTCATCTTCTTCTCGAAACCGTACGGACGACACGGAGAGCCCTACAGAGGATACGTACTAACCTTTTTCGTTTCCTTACTGTTTCTACTTATCGCGAATTTGAACGCGATTGCTCCGTTGATAACAAATTTTTACTTGGCCTCATACGCACTTATTAACTTTTGCACCTTTCACGCGGCATTCGTTCACCAAATTAATTGGCGACCCACGTTTCGCTATTACAACAAATGGCTATCGTTGGCCGGGTTCCTAATGTGTGCTTCAATAATGATGATAATCAGCTGGGCGATGGCACTGACTACAATGTTTGTTTTCATGACATTATATCTTCTCGTTCTTTATAGGAAGCCAGACGTGAGCTGGGGTAGCAGCACTGAAGGACAACGTTACCAGGACACGGTCTCATTTATAACTGAGCTCAGTCACCGAGTGACGCACAGTCGCTCGTACAACCCTCAGCTGCTTGTGCTCGCTGGGACACCGCTACAGCGCCCCGCGCTCATCGACGTAGCACACATGCTCACGAAGATAGGATCATTCATGATAGTTGCTGACATTACAGACACTGCATTAACCAAAGGCGAACGTGTCAGCCGTCAGCACATAGGCGAGCAGTTTGTGAAGTATCGCAAACTCCGCGCGTTCTACGTTCTCCTGCAAGGGCTTGACTTGAAGCGCGGCGCCAACGCACTGATGCAAGCTAGTGGGCTTGGAAAACTCTCTCCTAACATATTACTCATAGGATTCAAAAAGGACTGGACGAACGCTGAACATAAACAGATTAGCGACTACTACGGAGTTATCCAAGACGCATTCGATTTGAAGCTTGCGGTAACGATACTGCGAGTCCCCGGGACCTCGCAAACAGTACAGCGGTGGAGGTGGACACCCCGTGTGGTCTCGGAACCGAATAACTTCGAGCTCGTGATGCACACACAAGTACTCGCCCTCATGAACTCTGATTCAGACCTCGAAGACGACGACCAAGCAATCGACGATGATTCTGACAAACTTATCGACTCTGAAAACCAGCTGTCAAACGAAACTTCAAGTAGCGAACAGAATCCAGAATCCATCGGCGGAGAAGCATTACGATCTGCGAACTTTTCCCGGACTTCAAGACGGAAACTTTCAAACGGAGCTTTGTCATTCAGAGCTTTGTCGTTCAATTGCAAACAATACGTAGATGCTTGGTGGCTTTATGAGGATGGTGGTTTGAACATATTGCTTCCGTATATTATTGCACGTCGCGGCTACAAACACAAACTGCCCCTTAGGATATTCGCGATCACGAAAGAACAGCACAAAAGAGAGTTTGAAAATAAATGCATGGAAAGTTTATTGAAAAAATTTCGAATCGAGTACACGGAGCTGACACTGGTGCGAGGGATTGGGGACGCGCCACCCGAGTCGAGTTGGAAAATGTTCAACAACATCATAAACGATTTTAAGTCTGAGGAGAAAACTGATATTTTAACATCGGAAGCCGAGTTGAAGAGTCAGTACAGTAAAACGAGCAGGCATTTGCGACTACGAAACCTTCTGTTGCAACATTCGACAAAAGCCGCTATCGTCATCATTACCCTGCCATTGCCGAGAAAGGTGTCGGCATCGCTGTACATGGCGTGGCTAGAGGTCCTCAGCCGAGATTTGCCGCCCGTGTTATTCGTACGTGGCAACGACTCCTGTGTGTTGGACGCGTAA
Protein
MDVRLLGDCGKMVPKSMLRRRRSESVEIGNTNCEVNSEENESRSTREYSNGHLHPTPELTTYGGIKLGWIQGVLIPCLLNIWGVMLFLRIAWVVAQAGIGLSVIIVFLSAIVCVITTLSLSAICTNGQLQGGGVYYLVSRSLGAEFGASVGILFAFANAVAASMNTIGFCDSLNHLLKTHGLKIVDNDVNDIRIVGAVALLVMCVICAIGMDWETKAQNFLIVVIVVAIINYIVGALIGPSSDVERAKGFVGINLQTIRENWLPNFRYSEGREHDFFSVFAVYFPAVTGVQAGANICGDLKNPAVAIPKGTLLALAISVTSYFTMVILSGMGALRDASGNVANLTTAALESCKPHCGYGLHNNYAIMQLMSAWGPFIYAGCWAATLSTALTNLLSVPRLIQALGVDRIYPGLIFFSKPYGRHGEPYRGYVLTFFVSLLFLLIANLNAIAPLITNFYLASYALINFCTFHAAFVHQINWRPTFRYYNKWLSLAGFLMCASIMMIISWAMALTTMFVFMTLYLLVLYRKPDVSWGSSTEGQRYQDTVSFITELSHRVTHSRSYNPQLLVLAGTPLQRPALIDVAHMLTKIGSFMIVADITDTALTKGERVSRQHIGEQFVKYRKLRAFYVLLQGLDLKRGANALMQASGLGKLSPNILLIGFKKDWTNAEHKQISDYYGVIQDAFDLKLAVTILRVPGTSQTVQRWRWTPRVVSEPNNFELVMHTQVLALMNSDSDLEDDDQAIDDDSDKLIDSENQLSNETSSSEQNPESIGGEALRSANFSRTSRRKLSNGALSFRALSFNCKQYVDAWWLYEDGGLNILLPYIIARRGYKHKLPLRIFAITKEQHKREFENKCMESLLKKFRIEYTELTLVRGIGDAPPESSWKMFNNIINDFKSEEKTDILTSEAELKSQYSKTSRHLRLRNLLLQHSTKAAIVIITLPLPRKVSASLYMAWLEVLSRDLPPVLFVRGNDSCVLDA
Summary
Uniprot
A0A2A4JN03
A0A194Q257
A0A2A4JL60
A0A067QMC3
A0A1B0EWP1
Q16UI3
+ More
A0A1S4FNN9 A0A023EWB6 A0A182G664 A0A1Q3FSA3 E0VSF3 B0WKT5 A0A1B6EM82 B0WKT4 A0A1S4FNU9 Q16UI2 A0A146L8H8 A0A0A9YLQ8 B4GF84 U4U4I0 Q296F6 N6TV98 B4JHV1 B4KD91 A0A0M5IZZ9 B4LZB6 A0A182JS22 W8BU02 A0A0K8UVZ0 A0A034VJ51 B4NL26 A0A1I8M5P1 T1PDT6 A0A1Y0AWN1 A0A182M535 Q5TXE0 A0A2C9GV90 A0A182PIC3 A0A182YK80 A0A182TD45 A0A1J1ID56 A0A182Q954 A0A2M3ZF49 A0A2M3YXG8 A0A182IIF6 A0A2M3YXH5 A0A182LCN8 A7UR19 A0A182M4E3 A0A182LCP0 A0A182UGN1 A0A2M4CR16 W5JVP7 E1ZW64 A0A2C9GV83 A0A182JN28 A0A182V5N2 A0A182VQN7 A0A182J3J7 A0A084W3D0 A0A182FSH2 A0A182RHW2 A0A182IIF5 A0A1Q3FV52 T1PLE3 A0A0L0BWL3 A0A182TW57 A0A1S4GCA3 A0A2M3YYV0 A0A182V675 A0A2M3YXH2 A0A2M3ZY80 A0A182IUZ5 A0A2M3ZYG8 A0A182HL76 Q8T5I9 A0A2M4BB00 A0A2M4BCB9 A0A182WSC5 D2A527 A0A182QW38 A0A1I8NN16 A0A067QZ58 A0A034V8A2 A0A182N5V8 A0A0K8VSD2
A0A1S4FNN9 A0A023EWB6 A0A182G664 A0A1Q3FSA3 E0VSF3 B0WKT5 A0A1B6EM82 B0WKT4 A0A1S4FNU9 Q16UI2 A0A146L8H8 A0A0A9YLQ8 B4GF84 U4U4I0 Q296F6 N6TV98 B4JHV1 B4KD91 A0A0M5IZZ9 B4LZB6 A0A182JS22 W8BU02 A0A0K8UVZ0 A0A034VJ51 B4NL26 A0A1I8M5P1 T1PDT6 A0A1Y0AWN1 A0A182M535 Q5TXE0 A0A2C9GV90 A0A182PIC3 A0A182YK80 A0A182TD45 A0A1J1ID56 A0A182Q954 A0A2M3ZF49 A0A2M3YXG8 A0A182IIF6 A0A2M3YXH5 A0A182LCN8 A7UR19 A0A182M4E3 A0A182LCP0 A0A182UGN1 A0A2M4CR16 W5JVP7 E1ZW64 A0A2C9GV83 A0A182JN28 A0A182V5N2 A0A182VQN7 A0A182J3J7 A0A084W3D0 A0A182FSH2 A0A182RHW2 A0A182IIF5 A0A1Q3FV52 T1PLE3 A0A0L0BWL3 A0A182TW57 A0A1S4GCA3 A0A2M3YYV0 A0A182V675 A0A2M3YXH2 A0A2M3ZY80 A0A182IUZ5 A0A2M3ZYG8 A0A182HL76 Q8T5I9 A0A2M4BB00 A0A2M4BCB9 A0A182WSC5 D2A527 A0A182QW38 A0A1I8NN16 A0A067QZ58 A0A034V8A2 A0A182N5V8 A0A0K8VSD2
Pubmed
EMBL
NWSH01001068
PCG72813.1
KQ459564
KPI99652.1
PCG72815.1
KK853171
+ More
KDR10319.1 AJWK01006428 CH477621 EAT38198.1 GAPW01000083 JAC13515.1 JXUM01000651 JXUM01000652 JXUM01000653 JXUM01000654 JXUM01000655 JXUM01000656 JXUM01000657 JXUM01000658 JXUM01000659 JXUM01000660 KQ560109 KXJ84476.1 GFDL01004689 JAV30356.1 DS235750 EEB16309.1 DS231976 EDS30058.1 GECZ01030739 GECZ01015000 JAS39030.1 JAS54769.1 EDS30057.1 EAT38199.1 GDHC01015659 JAQ02970.1 GBHO01013124 JAG30480.1 CH479182 EDW34269.1 KB630198 KB631707 ERL83470.1 ERL85501.1 CM000070 EAL28501.1 APGK01010025 APGK01010026 APGK01010027 APGK01053218 APGK01053219 APGK01053220 KB741223 KB738096 ENN72321.1 ENN82770.1 CH916369 EDV92859.1 CH933806 EDW14873.1 CP012526 ALC46644.1 CH940650 EDW68151.1 GAMC01013964 GAMC01013961 JAB92594.1 GDHF01026214 GDHF01021482 GDHF01008314 JAI26100.1 JAI30832.1 JAI44000.1 GAKP01016429 GAKP01016423 JAC42529.1 CH964272 EDW84229.1 KA646937 AFP61566.1 KY921816 ART29405.1 AXCM01001650 AAAB01008794 EAL42059.4 CVRI01000047 CRK98147.1 AXCN02000174 GGFM01006391 MBW27142.1 GGFM01000175 MBW20926.1 APCN01002153 GGFM01000210 MBW20961.1 EDO64829.2 GGFL01003443 MBW67621.1 ADMH02000281 ETN67130.1 GL434776 EFN74573.1 ATLV01019917 ATLV01019918 KE525285 KFB44724.1 GFDL01003571 JAV31474.1 KA649571 AFP64200.1 JRES01001238 KNC24410.1 AAAB01008987 GGFM01000684 MBW21435.1 GGFM01000216 MBW20967.1 GGFK01000205 MBW33526.1 GGFK01000189 MBW33510.1 APCN01000883 AJ439353 CAD27923.1 EAA00828.4 GGFJ01001059 MBW50200.1 GGFJ01001558 MBW50699.1 KQ971361 EFA05305.2 AXCN02001150 KDR10326.1 GAKP01021184 JAC37768.1 GDHF01010819 GDHF01004981 JAI41495.1 JAI47333.1
KDR10319.1 AJWK01006428 CH477621 EAT38198.1 GAPW01000083 JAC13515.1 JXUM01000651 JXUM01000652 JXUM01000653 JXUM01000654 JXUM01000655 JXUM01000656 JXUM01000657 JXUM01000658 JXUM01000659 JXUM01000660 KQ560109 KXJ84476.1 GFDL01004689 JAV30356.1 DS235750 EEB16309.1 DS231976 EDS30058.1 GECZ01030739 GECZ01015000 JAS39030.1 JAS54769.1 EDS30057.1 EAT38199.1 GDHC01015659 JAQ02970.1 GBHO01013124 JAG30480.1 CH479182 EDW34269.1 KB630198 KB631707 ERL83470.1 ERL85501.1 CM000070 EAL28501.1 APGK01010025 APGK01010026 APGK01010027 APGK01053218 APGK01053219 APGK01053220 KB741223 KB738096 ENN72321.1 ENN82770.1 CH916369 EDV92859.1 CH933806 EDW14873.1 CP012526 ALC46644.1 CH940650 EDW68151.1 GAMC01013964 GAMC01013961 JAB92594.1 GDHF01026214 GDHF01021482 GDHF01008314 JAI26100.1 JAI30832.1 JAI44000.1 GAKP01016429 GAKP01016423 JAC42529.1 CH964272 EDW84229.1 KA646937 AFP61566.1 KY921816 ART29405.1 AXCM01001650 AAAB01008794 EAL42059.4 CVRI01000047 CRK98147.1 AXCN02000174 GGFM01006391 MBW27142.1 GGFM01000175 MBW20926.1 APCN01002153 GGFM01000210 MBW20961.1 EDO64829.2 GGFL01003443 MBW67621.1 ADMH02000281 ETN67130.1 GL434776 EFN74573.1 ATLV01019917 ATLV01019918 KE525285 KFB44724.1 GFDL01003571 JAV31474.1 KA649571 AFP64200.1 JRES01001238 KNC24410.1 AAAB01008987 GGFM01000684 MBW21435.1 GGFM01000216 MBW20967.1 GGFK01000205 MBW33526.1 GGFK01000189 MBW33510.1 APCN01000883 AJ439353 CAD27923.1 EAA00828.4 GGFJ01001059 MBW50200.1 GGFJ01001558 MBW50699.1 KQ971361 EFA05305.2 AXCN02001150 KDR10326.1 GAKP01021184 JAC37768.1 GDHF01010819 GDHF01004981 JAI41495.1 JAI47333.1
Proteomes
UP000218220
UP000053268
UP000027135
UP000092461
UP000008820
UP000069940
+ More
UP000249989 UP000009046 UP000002320 UP000008744 UP000030742 UP000001819 UP000019118 UP000001070 UP000009192 UP000092553 UP000008792 UP000075881 UP000007798 UP000095301 UP000075883 UP000007062 UP000075884 UP000075885 UP000076408 UP000075902 UP000183832 UP000075886 UP000075840 UP000075882 UP000000673 UP000000311 UP000075880 UP000075903 UP000075920 UP000030765 UP000069272 UP000075900 UP000037069 UP000076407 UP000007266 UP000095300
UP000249989 UP000009046 UP000002320 UP000008744 UP000030742 UP000001819 UP000019118 UP000001070 UP000009192 UP000092553 UP000008792 UP000075881 UP000007798 UP000095301 UP000075883 UP000007062 UP000075884 UP000075885 UP000076408 UP000075902 UP000183832 UP000075886 UP000075840 UP000075882 UP000000673 UP000000311 UP000075880 UP000075903 UP000075920 UP000030765 UP000069272 UP000075900 UP000037069 UP000076407 UP000007266 UP000095300
Interpro
SUPFAM
SSF81665
SSF81665
ProteinModelPortal
A0A2A4JN03
A0A194Q257
A0A2A4JL60
A0A067QMC3
A0A1B0EWP1
Q16UI3
+ More
A0A1S4FNN9 A0A023EWB6 A0A182G664 A0A1Q3FSA3 E0VSF3 B0WKT5 A0A1B6EM82 B0WKT4 A0A1S4FNU9 Q16UI2 A0A146L8H8 A0A0A9YLQ8 B4GF84 U4U4I0 Q296F6 N6TV98 B4JHV1 B4KD91 A0A0M5IZZ9 B4LZB6 A0A182JS22 W8BU02 A0A0K8UVZ0 A0A034VJ51 B4NL26 A0A1I8M5P1 T1PDT6 A0A1Y0AWN1 A0A182M535 Q5TXE0 A0A2C9GV90 A0A182PIC3 A0A182YK80 A0A182TD45 A0A1J1ID56 A0A182Q954 A0A2M3ZF49 A0A2M3YXG8 A0A182IIF6 A0A2M3YXH5 A0A182LCN8 A7UR19 A0A182M4E3 A0A182LCP0 A0A182UGN1 A0A2M4CR16 W5JVP7 E1ZW64 A0A2C9GV83 A0A182JN28 A0A182V5N2 A0A182VQN7 A0A182J3J7 A0A084W3D0 A0A182FSH2 A0A182RHW2 A0A182IIF5 A0A1Q3FV52 T1PLE3 A0A0L0BWL3 A0A182TW57 A0A1S4GCA3 A0A2M3YYV0 A0A182V675 A0A2M3YXH2 A0A2M3ZY80 A0A182IUZ5 A0A2M3ZYG8 A0A182HL76 Q8T5I9 A0A2M4BB00 A0A2M4BCB9 A0A182WSC5 D2A527 A0A182QW38 A0A1I8NN16 A0A067QZ58 A0A034V8A2 A0A182N5V8 A0A0K8VSD2
A0A1S4FNN9 A0A023EWB6 A0A182G664 A0A1Q3FSA3 E0VSF3 B0WKT5 A0A1B6EM82 B0WKT4 A0A1S4FNU9 Q16UI2 A0A146L8H8 A0A0A9YLQ8 B4GF84 U4U4I0 Q296F6 N6TV98 B4JHV1 B4KD91 A0A0M5IZZ9 B4LZB6 A0A182JS22 W8BU02 A0A0K8UVZ0 A0A034VJ51 B4NL26 A0A1I8M5P1 T1PDT6 A0A1Y0AWN1 A0A182M535 Q5TXE0 A0A2C9GV90 A0A182PIC3 A0A182YK80 A0A182TD45 A0A1J1ID56 A0A182Q954 A0A2M3ZF49 A0A2M3YXG8 A0A182IIF6 A0A2M3YXH5 A0A182LCN8 A7UR19 A0A182M4E3 A0A182LCP0 A0A182UGN1 A0A2M4CR16 W5JVP7 E1ZW64 A0A2C9GV83 A0A182JN28 A0A182V5N2 A0A182VQN7 A0A182J3J7 A0A084W3D0 A0A182FSH2 A0A182RHW2 A0A182IIF5 A0A1Q3FV52 T1PLE3 A0A0L0BWL3 A0A182TW57 A0A1S4GCA3 A0A2M3YYV0 A0A182V675 A0A2M3YXH2 A0A2M3ZY80 A0A182IUZ5 A0A2M3ZYG8 A0A182HL76 Q8T5I9 A0A2M4BB00 A0A2M4BCB9 A0A182WSC5 D2A527 A0A182QW38 A0A1I8NN16 A0A067QZ58 A0A034V8A2 A0A182N5V8 A0A0K8VSD2
Ontologies
GO
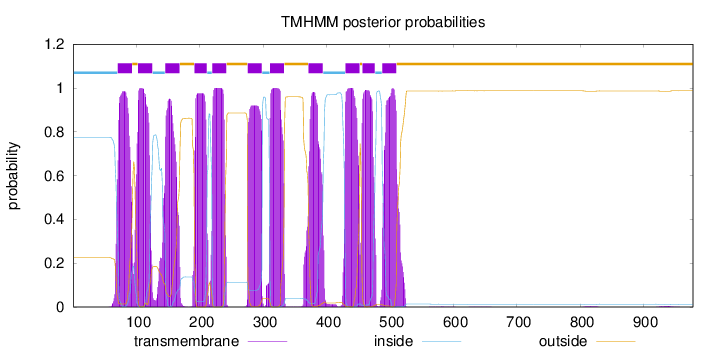
Topology
Length:
978
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
242.082000000001
Exp number, first 60 AAs:
0.0035
Total prob of N-in:
0.77390
inside
1 - 70
TMhelix
71 - 93
outside
94 - 102
TMhelix
103 - 125
inside
126 - 145
TMhelix
146 - 168
outside
169 - 191
TMhelix
192 - 211
inside
212 - 219
TMhelix
220 - 242
outside
243 - 275
TMhelix
276 - 298
inside
299 - 310
TMhelix
311 - 333
outside
334 - 371
TMhelix
372 - 394
inside
395 - 429
TMhelix
430 - 452
outside
453 - 456
TMhelix
457 - 476
inside
477 - 487
TMhelix
488 - 510
outside
511 - 978
Population Genetic Test Statistics
Pi
180.73743
Theta
146.316978
Tajima's D
0.50412
CLR
0.477912
CSRT
0.51632418379081
Interpretation
Uncertain
