Gene
KWMTBOMO12479
Annotation
PREDICTED:_tyrosine-protein_phosphatase_non-receptor_type_9_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.704
Sequence
CDS
ATGGCGGTGGCGCTGTCACCAGAACAAGAGCTGGCGACAAAAACATTTCTGAACGAGCTCAGCGAGACTGGTGCGGGGTGTGTATCACGAGCGACGGCGATCAAGTTCTTGTTGGCGCGCAAGTTCGATGTTGGGAGGGCCCACGCGCTCTGGAGGCAGCATGAGGCCACGCGTCGAAGGGAGGGACTCAACAAGTTTGAACCGTTCGAGGATCCACTTAAGTCAGAACTCGAAACCGGCAAATTTACTATACTGCCAACCAGAGATGCGACCGGTGCTGCCATTGCCGTGTTCACAGCGAACAAACACTTCCCCTCACAGACTACGCATCAAACAACACTACAGGGTGTGGTGTACCAGTTGGACGTGGCCTTACAATCGCTGGAGACTCAGCGAGCCGGCCTGGTCTTCATCTACGACATGACCGACTCCAAATACACCAACTTCGACTACGAGCTGTCGCAGAAAATATTGACTATGTTAAAGGGTGGTTATCCGGCCAAATTGAAGAAGGTATTAATAGTGACGGCGCCGCTTTGGTTCAAGGCTCCCTTCCGGATACTTCGGTTGTTCGTTAGAGAGAAGTTACGGGAACGCGTGCACACGGTTAGCGCGCCGCAACTTGGCGCGCACGTGCCGCGATCTAGCCTGCCCAAGCAACTGGGTGGCTTGCACGACACTGATCACACCACTTGGTTGGAATATTGCAAAAACTCTTACATAAACAACATTCAGAACTCTAAATTGGACGGCGTTATCGATGAATATGTAATCAGTAACCACATACCGCCCGTCGAACCGCAAAATGGTATAATAGAGCATCAGAGTGACATGACAAGCGATCGTGCAGTCAGACTCGGAGATACAGGGACCAAAATGCACATCAGCGGCGACCCGCCCTACACGTGCGACCCGAGCCCTTTGAGGCACTCGCATGGGTATAACGGCGATACTAAAAGCAGACACATTAACTCCGGCGACGAGGATGATATAGACATAAGCACGCGCGGCACGTGGTCGTCGGGCGAGCTGTCCCCCGGGCTGTCGGACGAGGAGTGCGCGGCGGGGGGCGCGGGCGGCGCGGGGGAGACGCCGGCGGCGCTGCTGGCCCGCCTGCGGCTCGTGCGGCGCCGGGGCCTCGTCGCAGAGTACGACGAGATACGGTCCCGCCCGCCCGTCGGAACCTTCCACAACGCCAAGTTGCCGAGCAACCTAGCCAAGAACCGCTACACGGACGTGCTGTGCTACGACCACTCGCGCGTGCTGTTGTCGCAGACCGACCCGGACGACCCCACCACGGACTACATCAACGCTAACTACGTCGACGGATACAAACAGAAGAACGCCTACATTTGTACACAGGGTCCTCTACCGAAGACGTTCGGTGACTTTTGGCGCATGGTGTGGGAGCAGGGCACCGTCGTGATAGTGATGACCACGCGCGTGATGGAGCGCGGGCGCGTCAAGTGCGGGCAGTACTGGCCCGCGGCGGCCGGCGCCAGGCAGCAGCACGGGCCCTACGCCGTGCTCGCCGAGGACGTGATCCAGCACGACGACTACACGATCAGCTCGCTGGTGCTGACGGAGCTGCGCACGGAGCAGGTGCGGCGCGTGTGGCACGGACAGTACACGCGCTGGCCGGACTACGGCGTGCCGGGCGGCGGCGGGGGCGGCGGCGGGGGCGGCGGGGCGGCGGGCGCGCGCGCGCTCCCAGTGCTGCGGTTCATGCAGCTCGTGCGCCGCGCGCAACAACGAGGACTCGCCGAGTTGGGAGACGCGTGGGCCGGACACAGCAAAGGGCCACCGATCGTCGTCCACTGCTCGGCCGGCATCGGACGGACAGGCACGTTCCTGACGCTGGACACGTGCGCGGCGCGGCTGGAGGCGGAGGGCGCGGCGGACGTGCGCGGCACGGTGGAGCGCATCCGGTCGCAGCGCGCGCACTCCATACAGATGCCCGACCAGTACGTGTTCTGTCATCTCGCGCTCCTCGAGTACGCGGTTATGAATGGATATTTAGAATCTGCCGATCTGACTGGCTTCGACGATGAGAACGATGAGGAGTCCGAATAA
Protein
MAVALSPEQELATKTFLNELSETGAGCVSRATAIKFLLARKFDVGRAHALWRQHEATRRREGLNKFEPFEDPLKSELETGKFTILPTRDATGAAIAVFTANKHFPSQTTHQTTLQGVVYQLDVALQSLETQRAGLVFIYDMTDSKYTNFDYELSQKILTMLKGGYPAKLKKVLIVTAPLWFKAPFRILRLFVREKLRERVHTVSAPQLGAHVPRSSLPKQLGGLHDTDHTTWLEYCKNSYINNIQNSKLDGVIDEYVISNHIPPVEPQNGIIEHQSDMTSDRAVRLGDTGTKMHISGDPPYTCDPSPLRHSHGYNGDTKSRHINSGDEDDIDISTRGTWSSGELSPGLSDEECAAGGAGGAGETPAALLARLRLVRRRGLVAEYDEIRSRPPVGTFHNAKLPSNLAKNRYTDVLCYDHSRVLLSQTDPDDPTTDYINANYVDGYKQKNAYICTQGPLPKTFGDFWRMVWEQGTVVIVMTTRVMERGRVKCGQYWPAAAGARQQHGPYAVLAEDVIQHDDYTISSLVLTELRTEQVRRVWHGQYTRWPDYGVPGGGGGGGGGGGAAGARALPVLRFMQLVRRAQQRGLAELGDAWAGHSKGPPIVVHCSAGIGRTGTFLTLDTCAARLEAEGAADVRGTVERIRSQRAHSIQMPDQYVFCHLALLEYAVMNGYLESADLTGFDDENDEESE
Summary
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
Uniprot
A0A3S2LMM0
A0A194Q063
A0A1L8DMM8
A0A1L8DMX5
A0A1B0CP45
A0A1L8DMR1
+ More
U5EPN3 Q179W1 A0A088AQ71 A0A195F3G6 A0A0C9RTF6 A0A026WAY4 E2B7H4 A0A310SJL6 F4W941 A0A158P041 A0A2M3Z3B2 A0A2M3ZGK3 A0A232FDL7 A0A0N0BHX1 A0A084W092 K7IME7 A0A0J7KSR1 A0A0L7RHM2 A0A3L8DQE7 A0A2A3E966 A0A154P747 A0A151WUB1 A0A195B5I7 A0A151ICX3 T1IWR5 A0A210Q2Y1 E0VJD3 K1QFA5 A0A1L8DL94 A0A1L8DL90 A0A1L8DLB1 A0A0C9R6R5 A0A1L8DLI3 A0A2M3Z4C3 A0A0C9QC45 E9I8J5 A0A195EDJ4 A0A2S2QVA3 A0A087TR08
U5EPN3 Q179W1 A0A088AQ71 A0A195F3G6 A0A0C9RTF6 A0A026WAY4 E2B7H4 A0A310SJL6 F4W941 A0A158P041 A0A2M3Z3B2 A0A2M3ZGK3 A0A232FDL7 A0A0N0BHX1 A0A084W092 K7IME7 A0A0J7KSR1 A0A0L7RHM2 A0A3L8DQE7 A0A2A3E966 A0A154P747 A0A151WUB1 A0A195B5I7 A0A151ICX3 T1IWR5 A0A210Q2Y1 E0VJD3 K1QFA5 A0A1L8DL94 A0A1L8DL90 A0A1L8DLB1 A0A0C9R6R5 A0A1L8DLI3 A0A2M3Z4C3 A0A0C9QC45 E9I8J5 A0A195EDJ4 A0A2S2QVA3 A0A087TR08
Pubmed
EMBL
RSAL01000055
RVE49971.1
KQ459585
KPI98384.1
GFDF01006382
JAV07702.1
+ More
GFDF01006384 JAV07700.1 AJWK01021415 AJWK01021416 AJWK01021417 AJWK01021418 AJWK01021419 AJWK01021420 AJWK01021421 AJWK01021422 GFDF01006383 JAV07701.1 GANO01000104 JAB59767.1 CH477344 EAT43037.1 KQ981855 KYN34926.1 GBYB01012030 JAG81797.1 KK107293 EZA53220.1 GL446181 EFN88349.1 KQ766694 OAD53592.1 GL888002 EGI69223.1 ADTU01000955 ADTU01000956 ADTU01000957 GGFM01002251 MBW23002.1 GGFM01006925 MBW27676.1 NNAY01000431 OXU28437.1 KQ435739 KOX76861.1 ATLV01019101 KE525262 KFB43636.1 LBMM01003563 KMQ93378.1 KQ414590 KOC70350.1 QOIP01000005 RLU22423.1 KZ288320 PBC28268.1 KQ434829 KZC07766.1 KQ982736 KYQ51443.1 KQ976598 KYM79540.1 KQ978002 KYM98210.1 JH431629 NEDP02005166 OWF43100.1 DS235222 EEB13489.1 JH817192 EKC27535.1 GFDF01006858 JAV07226.1 GFDF01006856 JAV07228.1 GFDF01006852 JAV07232.1 GBYB01012029 JAG81796.1 GFDF01006857 JAV07227.1 GGFM01002621 MBW23372.1 GBYB01012028 JAG81795.1 GL761587 EFZ23095.1 KQ979039 KYN23295.1 GGMS01012462 MBY81665.1 KK116364 KFM67547.1
GFDF01006384 JAV07700.1 AJWK01021415 AJWK01021416 AJWK01021417 AJWK01021418 AJWK01021419 AJWK01021420 AJWK01021421 AJWK01021422 GFDF01006383 JAV07701.1 GANO01000104 JAB59767.1 CH477344 EAT43037.1 KQ981855 KYN34926.1 GBYB01012030 JAG81797.1 KK107293 EZA53220.1 GL446181 EFN88349.1 KQ766694 OAD53592.1 GL888002 EGI69223.1 ADTU01000955 ADTU01000956 ADTU01000957 GGFM01002251 MBW23002.1 GGFM01006925 MBW27676.1 NNAY01000431 OXU28437.1 KQ435739 KOX76861.1 ATLV01019101 KE525262 KFB43636.1 LBMM01003563 KMQ93378.1 KQ414590 KOC70350.1 QOIP01000005 RLU22423.1 KZ288320 PBC28268.1 KQ434829 KZC07766.1 KQ982736 KYQ51443.1 KQ976598 KYM79540.1 KQ978002 KYM98210.1 JH431629 NEDP02005166 OWF43100.1 DS235222 EEB13489.1 JH817192 EKC27535.1 GFDF01006858 JAV07226.1 GFDF01006856 JAV07228.1 GFDF01006852 JAV07232.1 GBYB01012029 JAG81796.1 GFDF01006857 JAV07227.1 GGFM01002621 MBW23372.1 GBYB01012028 JAG81795.1 GL761587 EFZ23095.1 KQ979039 KYN23295.1 GGMS01012462 MBY81665.1 KK116364 KFM67547.1
Proteomes
UP000283053
UP000053268
UP000092461
UP000008820
UP000005203
UP000078541
+ More
UP000053097 UP000008237 UP000007755 UP000005205 UP000215335 UP000053105 UP000030765 UP000002358 UP000036403 UP000053825 UP000279307 UP000242457 UP000076502 UP000075809 UP000078540 UP000078542 UP000242188 UP000009046 UP000005408 UP000078492 UP000054359
UP000053097 UP000008237 UP000007755 UP000005205 UP000215335 UP000053105 UP000030765 UP000002358 UP000036403 UP000053825 UP000279307 UP000242457 UP000076502 UP000075809 UP000078540 UP000078542 UP000242188 UP000009046 UP000005408 UP000078492 UP000054359
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A3S2LMM0
A0A194Q063
A0A1L8DMM8
A0A1L8DMX5
A0A1B0CP45
A0A1L8DMR1
+ More
U5EPN3 Q179W1 A0A088AQ71 A0A195F3G6 A0A0C9RTF6 A0A026WAY4 E2B7H4 A0A310SJL6 F4W941 A0A158P041 A0A2M3Z3B2 A0A2M3ZGK3 A0A232FDL7 A0A0N0BHX1 A0A084W092 K7IME7 A0A0J7KSR1 A0A0L7RHM2 A0A3L8DQE7 A0A2A3E966 A0A154P747 A0A151WUB1 A0A195B5I7 A0A151ICX3 T1IWR5 A0A210Q2Y1 E0VJD3 K1QFA5 A0A1L8DL94 A0A1L8DL90 A0A1L8DLB1 A0A0C9R6R5 A0A1L8DLI3 A0A2M3Z4C3 A0A0C9QC45 E9I8J5 A0A195EDJ4 A0A2S2QVA3 A0A087TR08
U5EPN3 Q179W1 A0A088AQ71 A0A195F3G6 A0A0C9RTF6 A0A026WAY4 E2B7H4 A0A310SJL6 F4W941 A0A158P041 A0A2M3Z3B2 A0A2M3ZGK3 A0A232FDL7 A0A0N0BHX1 A0A084W092 K7IME7 A0A0J7KSR1 A0A0L7RHM2 A0A3L8DQE7 A0A2A3E966 A0A154P747 A0A151WUB1 A0A195B5I7 A0A151ICX3 T1IWR5 A0A210Q2Y1 E0VJD3 K1QFA5 A0A1L8DL94 A0A1L8DL90 A0A1L8DLB1 A0A0C9R6R5 A0A1L8DLI3 A0A2M3Z4C3 A0A0C9QC45 E9I8J5 A0A195EDJ4 A0A2S2QVA3 A0A087TR08
PDB
4ICZ
E-value=7.07394e-73,
Score=699
Ontologies
GO
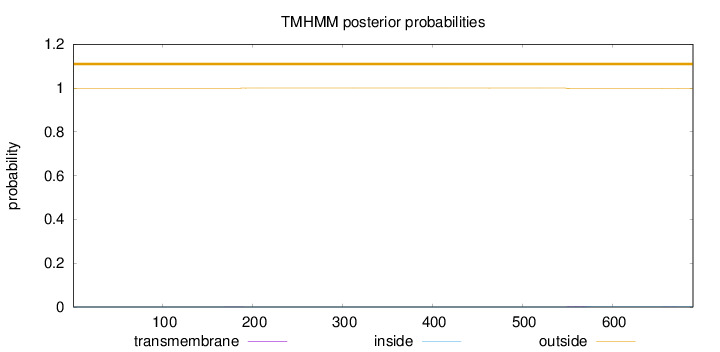
Topology
Length:
690
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0652600000000001
Exp number, first 60 AAs:
0.00082
Total prob of N-in:
0.00051
outside
1 - 690
Population Genetic Test Statistics
Pi
21.339613
Theta
165.872105
Tajima's D
-2.263363
CLR
1.731816
CSRT
0.00439978001099945
Interpretation
Possibly Positive selection
