Gene
KWMTBOMO12476
Pre Gene Modal
BGIBMGA001475
Annotation
PREDICTED:_uncharacterized_protein_LOC101744275_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.61
Sequence
CDS
ATGTACAACGACGAGGAAGACAACGAGAGATATTTTAGTCCAGATTTACTTCATGTCGTTCGCATTGCCACTATTTCAAATCTTGAACTAGCTCCGTACGGGATTAGGAGTGTACCCACAAAAACTGAAGATGGAATACCGATTAGAAAGCTTTACGTTACTAACTTACCACCCAAGACAACCAGAACTGAATTGTATGGGGTATTTGCCCAATATGGTCTCATTAAAAGCTGCTGGTTGAAAATGGGAGACAAGGGGCCAAATAAAACTCCAATACCCACATATGCATTTGTAACTTTTAGTAATCCAGCAGACGCTCATCAAGCCCTTGAAGCACCTAGTCATGAAAAAATATTGAGGGGTCGAAACCTATATACGTCACCAGCAGACAGTTGGCATCAGCCAGTAGAGGATGCAGATGGTCGGGTCTGCTGGAAACCACGTTTTTCACGTACTAATGTTCGATCACGCTCTAGGTCAAAAGATTTAGATGAAGCAGGTACAAGCACCAAAATGTCCACCGTTGCCACTTTAGATTCATCAGAATCATCAGATGAAGAAGATGTGAAGAGTTGCACCATATTAGATGTATTAAATAGAGATTGTTTGACGCATATCCTAAATTATATCCCTATTAGGGACATAATATTATCTGAAAGGGTCAGTAAAGGTTGGCAGATTATGGTCCAAGAATATTTAGCAGGCGTGCGCGTGCTGAAGACGTCGTGGTGGCAGGCGGAGGCGGCTCGGCTGACGACGGCGGTGCTGCGGCGGGTGCTGGCGCGCGTAGGCGACTCGCTGCTGGCGCTGCACATCGACCACCACTGGTCGGCGCTCAACGACCGCACGGCGCACTCCATCGCCAGGTTCTGTCCCAAGCTCGAGGAGCTCAAGATCGTCGGTATGCATACAAAAAATTGGAACCCCCTTCTGTACGGATGTAAAAAACTGAAGAATTTATCATTTATTTCTTGCAACAAGCTGACGGACTCGAGCCTGGTGCATCTGGCGCACGTGGACTCCTGCATCGAGAAGATCACCGTGGCCAACAACACGCACGTGACGGGCCTGTTCCTCACCGGCGCCGCCCCGCCGCAGCTGAGCTCGCTGTCCTTCTACAACTGCTACAGCCTGCAGGGGACGGTCCTAAGCGCCGCCATGGACTCCCTACCGAACCTGACCACCTTAAAGCTGGACCTGTGTCCAGCGGCCATGTGGAAAATTGTACCCATGATCTTAAAGACCCTATCCAAACTCGAGGAGCTGTCTCTGAGCGAGTACATGTCGAGCGAGGTGTGCTTCACGACCCAGAGCAGTGATGAATTCTGTGAAGCCCTCACCGGTCTCACCGAGCTACGGAGATTGAACTTGAGCAGAAACATACACATTACTAATGCAGTGCTCAAGCAAATCGGACAAACGTGTCATAAGTTAGAGTACCTCAACGTATCAAGTTGCAATTCCAGAAAAAACTTCCCTCAACCGGGAGTGAGCGACGAAGGTCTGGCGGCGCTGTGGGCGGGGTGCCGGCGGCTGCGCGAGCTCGACGTGTCGTACCTGGCGGGGCTGGGCGGCGCCGGGCTGGCGGGACAGGGCGGCGCCGCGCGGCTGCGTCGGTTCACGGCGCGCGGCAACCCCGCGCTGTCCGCCGCCGGGCCCGCCGCGCTGCTCGCCGCCGCGCCCTGTCTGCAGGAGATGGATCTGTGCGGGTGCGACGGAGTGTCCGAAGAAATAGTGGAGGCCGCAGTGAACGCCCTGGCGCTGAACCCGCGACACCTGCTCCTGCGGCTCGCGGGCACCGGCGCCGCGCAGGCACAGGAGGAGCTGCCGAAGCACAGGCTGCTGACGGTGAACGTGGCGGACGACCTGTGCAACCCCAACATGAGGCCGGACTTCGTGGACCGCATATTCGGGAACGACTCCGACGAGTCCTTCGACGACCTCTACGACCACGACGATTTCGAGGACCTGTTCGGTTCGGACGAGGAAATATTCTTAGAGGACGAGCTAGACGAATACGAGCAGATCTTCGCGTACGGAATCGATGTCCCCGACATACTCATATAG
Protein
MYNDEEDNERYFSPDLLHVVRIATISNLELAPYGIRSVPTKTEDGIPIRKLYVTNLPPKTTRTELYGVFAQYGLIKSCWLKMGDKGPNKTPIPTYAFVTFSNPADAHQALEAPSHEKILRGRNLYTSPADSWHQPVEDADGRVCWKPRFSRTNVRSRSRSKDLDEAGTSTKMSTVATLDSSESSDEEDVKSCTILDVLNRDCLTHILNYIPIRDIILSERVSKGWQIMVQEYLAGVRVLKTSWWQAEAARLTTAVLRRVLARVGDSLLALHIDHHWSALNDRTAHSIARFCPKLEELKIVGMHTKNWNPLLYGCKKLKNLSFISCNKLTDSSLVHLAHVDSCIEKITVANNTHVTGLFLTGAAPPQLSSLSFYNCYSLQGTVLSAAMDSLPNLTTLKLDLCPAAMWKIVPMILKTLSKLEELSLSEYMSSEVCFTTQSSDEFCEALTGLTELRRLNLSRNIHITNAVLKQIGQTCHKLEYLNVSSCNSRKNFPQPGVSDEGLAALWAGCRRLRELDVSYLAGLGGAGLAGQGGAARLRRFTARGNPALSAAGPAALLAAAPCLQEMDLCGCDGVSEEIVEAAVNALALNPRHLLLRLAGTGAAQAQEELPKHRLLTVNVADDLCNPNMRPDFVDRIFGNDSDESFDDLYDHDDFEDLFGSDEEIFLEDELDEYEQIFAYGIDVPDILI
Summary
Uniprot
H9IW44
A0A2A4JL70
A0A212FL60
A0A2A4JMK9
A0A2A4JN13
A0A194Q058
+ More
A0A194RPY0 A0A3S2TMA6 A0A1B6LI15 A0A1B6LUX8 A0A1B6LC54 A0A1B6MFA5 A0A1B6HBQ6 A0A1B6IBN6 A0A1B6ITP1 A0A1B6J5K8 A0A1B6F954 A0A1B6H144 A0A2J7PZU4 A0A026VWA8 A0A3L8DUR9 A0A151JYS8 F4X2G1 A0A158NW27 A0A232FAK1 A0A154PGR8 A0A195E2X0 A0A195CSP2 A0A087ZQJ4 A0A069DVE1 A0A0V0G3A9 A0A2A3E5X4 A0A224XLP0 A0A1B6CYQ3 A0A0A9ZEI6 A0A146M8R7 A0A0A9ZAM8 A0A023G8F5 K7JGG0 A0A1E1XB14 A0A1B0CDB6 L7LQ87 A0A131YWF5 V5II66 V5HWM2 A0A224YWB6 A0A131X980 A0A3M6TJT3 A0A2B4S849 T1PGM2
A0A194RPY0 A0A3S2TMA6 A0A1B6LI15 A0A1B6LUX8 A0A1B6LC54 A0A1B6MFA5 A0A1B6HBQ6 A0A1B6IBN6 A0A1B6ITP1 A0A1B6J5K8 A0A1B6F954 A0A1B6H144 A0A2J7PZU4 A0A026VWA8 A0A3L8DUR9 A0A151JYS8 F4X2G1 A0A158NW27 A0A232FAK1 A0A154PGR8 A0A195E2X0 A0A195CSP2 A0A087ZQJ4 A0A069DVE1 A0A0V0G3A9 A0A2A3E5X4 A0A224XLP0 A0A1B6CYQ3 A0A0A9ZEI6 A0A146M8R7 A0A0A9ZAM8 A0A023G8F5 K7JGG0 A0A1E1XB14 A0A1B0CDB6 L7LQ87 A0A131YWF5 V5II66 V5HWM2 A0A224YWB6 A0A131X980 A0A3M6TJT3 A0A2B4S849 T1PGM2
Pubmed
EMBL
BABH01005893
BABH01005894
BABH01005895
BABH01005896
BABH01005897
NWSH01001068
+ More
PCG72825.1 AGBW02007840 OWR54483.1 PCG72824.1 PCG72823.1 KQ459585 KPI98379.1 KQ459896 KPJ19370.1 RSAL01000055 RVE49967.1 GEBQ01016636 JAT23341.1 GEBQ01012527 JAT27450.1 GEBQ01018726 JAT21251.1 GEBQ01005340 JAT34637.1 GECU01035647 JAS72059.1 GECU01023369 JAS84337.1 GECU01017407 JAS90299.1 GECU01013248 JAS94458.1 GECZ01023078 JAS46691.1 GECZ01001402 JAS68367.1 NEVH01020332 PNF21851.1 KK107785 EZA47801.1 QOIP01000004 RLU24121.1 KQ981477 KYN41465.1 GL888578 EGI59386.1 ADTU01027780 NNAY01000522 OXU27874.1 KQ434900 KZC11051.1 KQ979701 KYN19493.1 KQ977329 KYN03547.1 GBGD01000831 JAC88058.1 GECL01003537 JAP02587.1 KZ288386 PBC26461.1 GFTR01007485 JAW08941.1 GEDC01018780 JAS18518.1 GBHO01001283 JAG42321.1 GDHC01003463 JAQ15166.1 GBHO01001282 GBRD01002119 GDHC01002606 JAG42322.1 JAG63702.1 JAQ16023.1 GBBM01006218 JAC29200.1 GFAC01002760 JAT96428.1 AJWK01007642 GACK01011219 JAA53815.1 GEDV01005579 JAP82978.1 GANP01003722 JAB80746.1 GANP01003721 JAB80747.1 GFPF01007118 MAA18264.1 GEFH01004487 JAP64094.1 RCHS01003465 RMX41645.1 LSMT01000169 PFX24702.1 KA647008 AFP61637.1
PCG72825.1 AGBW02007840 OWR54483.1 PCG72824.1 PCG72823.1 KQ459585 KPI98379.1 KQ459896 KPJ19370.1 RSAL01000055 RVE49967.1 GEBQ01016636 JAT23341.1 GEBQ01012527 JAT27450.1 GEBQ01018726 JAT21251.1 GEBQ01005340 JAT34637.1 GECU01035647 JAS72059.1 GECU01023369 JAS84337.1 GECU01017407 JAS90299.1 GECU01013248 JAS94458.1 GECZ01023078 JAS46691.1 GECZ01001402 JAS68367.1 NEVH01020332 PNF21851.1 KK107785 EZA47801.1 QOIP01000004 RLU24121.1 KQ981477 KYN41465.1 GL888578 EGI59386.1 ADTU01027780 NNAY01000522 OXU27874.1 KQ434900 KZC11051.1 KQ979701 KYN19493.1 KQ977329 KYN03547.1 GBGD01000831 JAC88058.1 GECL01003537 JAP02587.1 KZ288386 PBC26461.1 GFTR01007485 JAW08941.1 GEDC01018780 JAS18518.1 GBHO01001283 JAG42321.1 GDHC01003463 JAQ15166.1 GBHO01001282 GBRD01002119 GDHC01002606 JAG42322.1 JAG63702.1 JAQ16023.1 GBBM01006218 JAC29200.1 GFAC01002760 JAT96428.1 AJWK01007642 GACK01011219 JAA53815.1 GEDV01005579 JAP82978.1 GANP01003722 JAB80746.1 GANP01003721 JAB80747.1 GFPF01007118 MAA18264.1 GEFH01004487 JAP64094.1 RCHS01003465 RMX41645.1 LSMT01000169 PFX24702.1 KA647008 AFP61637.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9IW44
A0A2A4JL70
A0A212FL60
A0A2A4JMK9
A0A2A4JN13
A0A194Q058
+ More
A0A194RPY0 A0A3S2TMA6 A0A1B6LI15 A0A1B6LUX8 A0A1B6LC54 A0A1B6MFA5 A0A1B6HBQ6 A0A1B6IBN6 A0A1B6ITP1 A0A1B6J5K8 A0A1B6F954 A0A1B6H144 A0A2J7PZU4 A0A026VWA8 A0A3L8DUR9 A0A151JYS8 F4X2G1 A0A158NW27 A0A232FAK1 A0A154PGR8 A0A195E2X0 A0A195CSP2 A0A087ZQJ4 A0A069DVE1 A0A0V0G3A9 A0A2A3E5X4 A0A224XLP0 A0A1B6CYQ3 A0A0A9ZEI6 A0A146M8R7 A0A0A9ZAM8 A0A023G8F5 K7JGG0 A0A1E1XB14 A0A1B0CDB6 L7LQ87 A0A131YWF5 V5II66 V5HWM2 A0A224YWB6 A0A131X980 A0A3M6TJT3 A0A2B4S849 T1PGM2
A0A194RPY0 A0A3S2TMA6 A0A1B6LI15 A0A1B6LUX8 A0A1B6LC54 A0A1B6MFA5 A0A1B6HBQ6 A0A1B6IBN6 A0A1B6ITP1 A0A1B6J5K8 A0A1B6F954 A0A1B6H144 A0A2J7PZU4 A0A026VWA8 A0A3L8DUR9 A0A151JYS8 F4X2G1 A0A158NW27 A0A232FAK1 A0A154PGR8 A0A195E2X0 A0A195CSP2 A0A087ZQJ4 A0A069DVE1 A0A0V0G3A9 A0A2A3E5X4 A0A224XLP0 A0A1B6CYQ3 A0A0A9ZEI6 A0A146M8R7 A0A0A9ZAM8 A0A023G8F5 K7JGG0 A0A1E1XB14 A0A1B0CDB6 L7LQ87 A0A131YWF5 V5II66 V5HWM2 A0A224YWB6 A0A131X980 A0A3M6TJT3 A0A2B4S849 T1PGM2
PDB
1X4A
E-value=0.000701398,
Score=104
Ontologies
GO
Topology
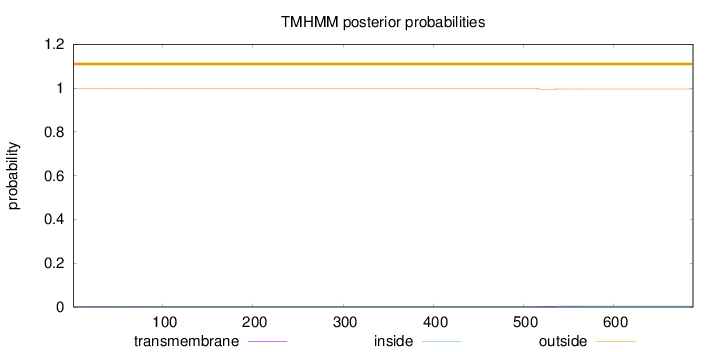
Length:
688
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11198
Exp number, first 60 AAs:
0.01082
Total prob of N-in:
0.00216
outside
1 - 688
Population Genetic Test Statistics
Pi
174.924885
Theta
179.054371
Tajima's D
-1.365482
CLR
143.701509
CSRT
0.0826458677066147
Interpretation
Uncertain
