Gene
KWMTBOMO12473 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001495
Annotation
PREDICTED:_ubiquitin-like_protein_7_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.198
Sequence
CDS
ATGGATAACCAGCCATGCATATTTTTGGGTATAAAAGTGAAACCTGGACCAATAGAACGTCACAAACTAGAGAATTTTGGACTCGAGAATACAGTAGATTCTCTTAGAACTGAGGCTGAAAAGCGATCCAACGTACCATCATCGTCATTAGAATTAATTCACCACGGAAAAATCTTAAAAGACAAGGCAACACTTCAAGAATGTGGAGTCAAGTCTGGTGAAATGGTGCATGTTGTGAAAAAGAAGACTACCACTGCCAGTGCAGCACCTCCAACTTACACTGATGCTGAGCTGCAGCAGTTAAATGCATCACTGAGAACTCTTGGATGTACACCAAATGCACCAGGTTGGACTAGAGCTATGCAGCTCTTAAATGATGACTCTGTTATGGCTGAGATCATAGAACAGGCCCCATCACTAGGAGAGGATTGTGTAGCTCTTTCAATATTGCACGAGGTTGAGTTATTGGCAGCTCTGGGTGCCAATCTACAAACAATGAGGCGGGGTGCCACAGCCCATCCAGAATTGCCCATAGCATTGAGACACCTCACAAATCTTGTTAGATCACGCTCTTCAACCGGATCTACTCCTGGTGATACTGCACCCACATCTGGATTTGCATATTCATTGGAGGCTTTGTCTGAAGATGAAGATGGTGAAGATGAGGAGACTGATGAAAATGGTGAAAGAAACCAAATAACTCAAGAACAGTTAGCAGCAGCATTGCAGGCGGCCACGGAAGCCGTGTCGACTCCGCGGAGCGGCGGCGGCAGTCACGAGAGCCTGCTGCGGTTGCTGCACGCCGGCGACTCCTCCCGACCGCACTCCTCCACGCAGACCTCAGGCCCCAGTCGCTCGGTTATCACTCCGGAAATGTTCAGTGAAGCTATTTCAGAAGCTATAAACAGATCCAGCAATAGACAAAGTGATGGAACACCTATGGAACAAAGTACTGCAGCATCTTCATCTCAAAGTCCCGGAGGAAGGGAAGATAATTTTACAACACAATTGAACCATATGCATGAAATGGGTCTACTGGACGACGCCCTAAATGTTCGCGCCCTACTAATATGCTCCGGTGACGTGAATGCAGCTATTAATTTAGTTTTTAGTGGCGCCATCGGAGATGAATAA
Protein
MDNQPCIFLGIKVKPGPIERHKLENFGLENTVDSLRTEAEKRSNVPSSSLELIHHGKILKDKATLQECGVKSGEMVHVVKKKTTTASAAPPTYTDAELQQLNASLRTLGCTPNAPGWTRAMQLLNDDSVMAEIIEQAPSLGEDCVALSILHEVELLAALGANLQTMRRGATAHPELPIALRHLTNLVRSRSSTGSTPGDTAPTSGFAYSLEALSEDEDGEDEETDENGERNQITQEQLAAALQAATEAVSTPRSGGGSHESLLRLLHAGDSSRPHSSTQTSGPSRSVITPEMFSEAISEAINRSSNRQSDGTPMEQSTAASSSQSPGGREDNFTTQLNHMHEMGLLDDALNVRALLICSGDVNAAINLVFSGAIGDE
Summary
Uniprot
H9IW64
A0A2H1W957
A0A2A4JN42
A0A194PYG3
A0A194RPD0
A0A212FL75
+ More
S4PIP0 A0A067R792 A0A1Y1NAP3 A0A0A9YLF5 A0A1W4X180 A0A0T6B6V8 E0VJF2 A0A195FXP0 D6W9J9 A0A195DS17 A0A195BS84 A0A158NVA5 F4X2L0 A0A154PEM4 A0A026W966 A0A151IBE2 A0A151WU32 V5G9F3 B0WFI3 A0A0N0BDD9 K7IME5 A0A0J7L6P0 A0A1Q3F451 A0A232FD72 A0A182GX56 Q16N61 U5ES39 A0A1E1XL41 A0A210QBP9 T1J358 A0A164LKQ3 A0A0P5HLD2 A0A0P6B2B4 A0A1S3HUN9 A0A0P5C4A0 A0A0P5WJ50 A0A0P4YJC4 A0A1S3HDQ1
S4PIP0 A0A067R792 A0A1Y1NAP3 A0A0A9YLF5 A0A1W4X180 A0A0T6B6V8 E0VJF2 A0A195FXP0 D6W9J9 A0A195DS17 A0A195BS84 A0A158NVA5 F4X2L0 A0A154PEM4 A0A026W966 A0A151IBE2 A0A151WU32 V5G9F3 B0WFI3 A0A0N0BDD9 K7IME5 A0A0J7L6P0 A0A1Q3F451 A0A232FD72 A0A182GX56 Q16N61 U5ES39 A0A1E1XL41 A0A210QBP9 T1J358 A0A164LKQ3 A0A0P5HLD2 A0A0P6B2B4 A0A1S3HUN9 A0A0P5C4A0 A0A0P5WJ50 A0A0P4YJC4 A0A1S3HDQ1
Pubmed
EMBL
BABH01005889
ODYU01007046
SOQ49476.1
NWSH01001068
PCG72820.1
KQ459585
+ More
KPI98376.1 KQ459896 KPJ19367.1 AGBW02007840 OWR54486.1 GAIX01000129 JAA92431.1 KK852693 KDR18317.1 GEZM01011984 JAV93316.1 GBHO01011671 GBHO01011668 GBRD01007471 GDHC01012104 GDHC01001435 JAG31933.1 JAG31936.1 JAG58350.1 JAQ06525.1 JAQ17194.1 LJIG01009479 KRT82997.1 DS235222 EEB13508.1 KQ981193 KYN45203.1 KQ971312 EEZ98142.1 KQ980530 KYN15607.1 KQ976417 KYM89880.1 ADTU01027051 GL888584 EGI59335.1 KQ434889 KZC10309.1 KK107324 QOIP01000006 EZA52533.1 RLU21507.1 KQ978106 KYM96975.1 KQ982745 KYQ51344.1 GALX01001696 JAB66770.1 DS231918 EDS26251.1 KQ435878 KOX69964.1 LBMM01000512 KMQ98174.1 GFDL01012728 JAV22317.1 NNAY01000431 OXU28443.1 JXUM01019912 KQ560558 KXJ81898.1 CH477837 EAT35781.1 GANO01002536 JAB57335.1 GFAA01003374 JAU00061.1 NEDP02004255 OWF46156.1 JH431820 LRGB01003123 KZS04207.1 GDIQ01227863 JAK23862.1 GDIP01147856 GDIP01023136 JAM80579.1 GDIP01180453 JAJ42949.1 GDIP01085601 JAM18114.1 GDIP01227943 JAI95458.1
KPI98376.1 KQ459896 KPJ19367.1 AGBW02007840 OWR54486.1 GAIX01000129 JAA92431.1 KK852693 KDR18317.1 GEZM01011984 JAV93316.1 GBHO01011671 GBHO01011668 GBRD01007471 GDHC01012104 GDHC01001435 JAG31933.1 JAG31936.1 JAG58350.1 JAQ06525.1 JAQ17194.1 LJIG01009479 KRT82997.1 DS235222 EEB13508.1 KQ981193 KYN45203.1 KQ971312 EEZ98142.1 KQ980530 KYN15607.1 KQ976417 KYM89880.1 ADTU01027051 GL888584 EGI59335.1 KQ434889 KZC10309.1 KK107324 QOIP01000006 EZA52533.1 RLU21507.1 KQ978106 KYM96975.1 KQ982745 KYQ51344.1 GALX01001696 JAB66770.1 DS231918 EDS26251.1 KQ435878 KOX69964.1 LBMM01000512 KMQ98174.1 GFDL01012728 JAV22317.1 NNAY01000431 OXU28443.1 JXUM01019912 KQ560558 KXJ81898.1 CH477837 EAT35781.1 GANO01002536 JAB57335.1 GFAA01003374 JAU00061.1 NEDP02004255 OWF46156.1 JH431820 LRGB01003123 KZS04207.1 GDIQ01227863 JAK23862.1 GDIP01147856 GDIP01023136 JAM80579.1 GDIP01180453 JAJ42949.1 GDIP01085601 JAM18114.1 GDIP01227943 JAI95458.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000027135
+ More
UP000192223 UP000009046 UP000078541 UP000007266 UP000078492 UP000078540 UP000005205 UP000007755 UP000076502 UP000053097 UP000279307 UP000078542 UP000075809 UP000002320 UP000053105 UP000002358 UP000036403 UP000215335 UP000069940 UP000249989 UP000008820 UP000242188 UP000076858 UP000085678
UP000192223 UP000009046 UP000078541 UP000007266 UP000078492 UP000078540 UP000005205 UP000007755 UP000076502 UP000053097 UP000279307 UP000078542 UP000075809 UP000002320 UP000053105 UP000002358 UP000036403 UP000215335 UP000069940 UP000249989 UP000008820 UP000242188 UP000076858 UP000085678
PRIDE
Interpro
ProteinModelPortal
H9IW64
A0A2H1W957
A0A2A4JN42
A0A194PYG3
A0A194RPD0
A0A212FL75
+ More
S4PIP0 A0A067R792 A0A1Y1NAP3 A0A0A9YLF5 A0A1W4X180 A0A0T6B6V8 E0VJF2 A0A195FXP0 D6W9J9 A0A195DS17 A0A195BS84 A0A158NVA5 F4X2L0 A0A154PEM4 A0A026W966 A0A151IBE2 A0A151WU32 V5G9F3 B0WFI3 A0A0N0BDD9 K7IME5 A0A0J7L6P0 A0A1Q3F451 A0A232FD72 A0A182GX56 Q16N61 U5ES39 A0A1E1XL41 A0A210QBP9 T1J358 A0A164LKQ3 A0A0P5HLD2 A0A0P6B2B4 A0A1S3HUN9 A0A0P5C4A0 A0A0P5WJ50 A0A0P4YJC4 A0A1S3HDQ1
S4PIP0 A0A067R792 A0A1Y1NAP3 A0A0A9YLF5 A0A1W4X180 A0A0T6B6V8 E0VJF2 A0A195FXP0 D6W9J9 A0A195DS17 A0A195BS84 A0A158NVA5 F4X2L0 A0A154PEM4 A0A026W966 A0A151IBE2 A0A151WU32 V5G9F3 B0WFI3 A0A0N0BDD9 K7IME5 A0A0J7L6P0 A0A1Q3F451 A0A232FD72 A0A182GX56 Q16N61 U5ES39 A0A1E1XL41 A0A210QBP9 T1J358 A0A164LKQ3 A0A0P5HLD2 A0A0P6B2B4 A0A1S3HUN9 A0A0P5C4A0 A0A0P5WJ50 A0A0P4YJC4 A0A1S3HDQ1
PDB
2DEN
E-value=0.00527421,
Score=94
Ontologies
GO
PANTHER
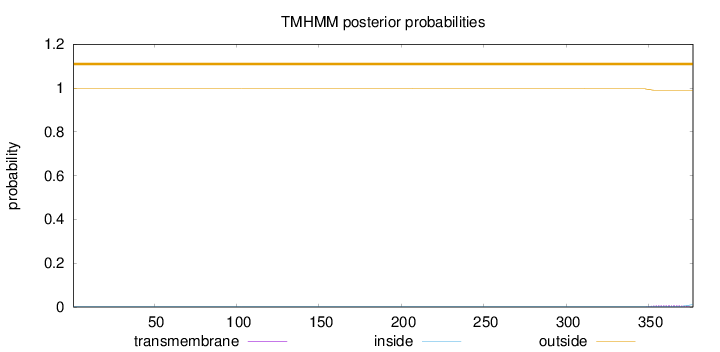
Topology
Length:
377
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15963
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00366
outside
1 - 377
Population Genetic Test Statistics
Pi
132.463154
Theta
162.820525
Tajima's D
-0.921126
CLR
0.156084
CSRT
0.148892555372231
Interpretation
Uncertain
