Gene
KWMTBOMO12469
Pre Gene Modal
BGIBMGA001494
Annotation
PREDICTED:_solute_carrier_family_25_member_46-A-like_isoform_X1_[Bombyx_mori]
Full name
Lysine--tRNA ligase
Alternative Name
Lysyl-tRNA synthetase
Location in the cell
PlasmaMembrane Reliability : 3.324
Sequence
CDS
ATGGCAGGTTTCGGAGATCATTCCGGTTATAACCGACCGTTTGGTTTAGATCCTAATCGAGAAACGAATGATTTATACGATACCCGTTACCAGCAGATTTATGGGTACCATACTCCAATTACTGAAGAGTTTCAAGGACCACCTCTTGTTATGGATGAAAACCCAGATGAAGATAACAATGCTTATATAACAACCGCAATAAGTGTAGCAAGTCTTTTAACAGAGAATTTGTTAAGTCATCCATTCATTGTACTCAGAAGACAATGTCAGGTCCATAACAATCTAAGGAACTATCACATAGTTCCTTACACTCTGTTACCTGTAGTAGTGAGACTACACAGGAGACAAGATGTCACAACTCTCTGGAAAGGGATTGGCTCCACTTGCATTGTGAGGGGTTTGAATTTAGCCGTTGAAGATGCTGTAGCCAAGGGCACTGGTTGGCCTAAGGAGGTGAACAAGTGCAATTCTGTTTTGCAATTTGTCCAGCACATAGCTCTAAAAAGCCTAAGTTTGGCATTAATAACTCCATTCTATTCTGCGAGCTTGGTAGAAACAGTGCAGAGTGATATCGCCAGTGACAAACCAGCTCTATTTGATGTGTTCAGAGAGGGATTTGTTCGTATACTCGAGTGGGGCACACCCAGTAAAGGACGCATGTTACCAATATGGGCAATTGTACTACCTACTGTAGCCCTAGGCGTTTCTAAATATATCTCACATATTACACTGAGAAGTTTAACTGCAAGAATACTCCGGTTCCAACAACGGCATAGGCATGAGCTGAGCGATTCATACAATTACAGCAAAAATGTACAGAATCCTCTTATAGAAGATATCAATCTGAAAGCAAACATTTTCTCATTTATTACTATGGAAATACTTTTTTATCCAGTGGAGACTATAATACATAGACTTCATATACAGGGTACTAGAACAATCATTGATAATTTAGACACAGGAACATCAGTGACACCTATTCTAACAGGATATGAAGGTTTCATGGACTGTTATAATACAACAATAGCCCGGGAAGGAATTTCAGGGTTGTACAAAGGGTTTGGTGCGCTAGTACTGCAGCTTGTAGCTCATATTTCAATAATTAAACTCACTACAATTGTAGTTACTGAAATAACGAACCTGTTGAAGCCTGTCAAGTCTCCTACAAACTCTGATGATCATACACAACAAAAGTACTTGTTTAATTAG
Protein
MAGFGDHSGYNRPFGLDPNRETNDLYDTRYQQIYGYHTPITEEFQGPPLVMDENPDEDNNAYITTAISVASLLTENLLSHPFIVLRRQCQVHNNLRNYHIVPYTLLPVVVRLHRRQDVTTLWKGIGSTCIVRGLNLAVEDAVAKGTGWPKEVNKCNSVLQFVQHIALKSLSLALITPFYSASLVETVQSDIASDKPALFDVFREGFVRILEWGTPSKGRMLPIWAIVLPTVALGVSKYISHITLRSLTARILRFQQRHRHELSDSYNYSKNVQNPLIEDINLKANIFSFITMEILFYPVETIIHRLHIQGTRTIIDNLDTGTSVTPILTGYEGFMDCYNTTIAREGISGLYKGFGALVLQLVAHISIIKLTTIVVTEITNLLKPVKSPTNSDDHTQQKYLFN
Summary
Catalytic Activity
ATP + L-lysine + tRNA(Lys) = AMP + diphosphate + L-lysyl-tRNA(Lys)
Similarity
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Uniprot
A0A3S2PHC9
A0A212FLC2
S4PS34
A0A2H1W9F0
A0A2A4JM08
A0A194PZA2
+ More
A0A194RPN1 A0A2P8ZC01 A0A0T6BFM2 Q16XZ0 A0A1S4FK72 A0A1Q3G2F2 A0A182HS89 A0A023F9P1 A0A182V973 A0A182LE70 Q7PQY0 U5ERT7 A0A182XAF0 A0A182U9G6 A0A182RTI5 A0A182PL56 A0A2M4BPF6 A0A182LV47 A0A182NR60 A0A2M4AI87 A0A2M3Z8A0 A0A2M4BNR0 A0A2M4BNR5 A0A2M4AHR6 A0A2M4AHW7 N6TKY0 T1HS92 A0A0P4VLB5 A0A182K3D9 W5JQ94 A0A224XRE7 A0A0K8TMY9 A0A182Y414 A0A0V0G7A8 A0A069DT22 A0A0L0CIM7 R4FKT1 A0A1A9UQB6 A0A1B0GA09 A0A0K8V3K1 A0A182QXK9 A0A1A9X8S6 A0A1B0C3A9 A0A034WLM3 A0A0A1XR71 A0A1A9ZEH7 W8CDN6 B4PXA5 A0A1I8NH60 A0A1I8NH52 Q9VXK8 A0A1I8PSJ5 A0A1I8PSG3 B4JX37 A0A1J1J6D2 B3NXG3 A0A336KEK8 A0A182VZV3 B4L6Y3 B4R5K6 A0A1W4VG58 A0A067QQG7 A0A0M4EN53 A0A0A9X8A3 B4MJ77 B3MPW7 A0A1B6DH59 A0A0P9AXA5 B4MA55 A0A0P8YKV4 A0A2R7WJ47 B4HCQ9 D6W807 A0A3B0K191 A0A087ZU62 A0A182GYW9 A0A1B6I2G2 E0VRK5 E2BNR4 A0A0N0BHY1 A0A1Y1LJK4 B0W6T3 A0A084VB74 A0A182FPH8 A0A2A3EH13 A0A1J1J949 E2AE71 A0A151I9T7 A0A026WGK3 A0A151HYQ5 A0A1W4WQ62 F4X481 A0A158N9I2
A0A194RPN1 A0A2P8ZC01 A0A0T6BFM2 Q16XZ0 A0A1S4FK72 A0A1Q3G2F2 A0A182HS89 A0A023F9P1 A0A182V973 A0A182LE70 Q7PQY0 U5ERT7 A0A182XAF0 A0A182U9G6 A0A182RTI5 A0A182PL56 A0A2M4BPF6 A0A182LV47 A0A182NR60 A0A2M4AI87 A0A2M3Z8A0 A0A2M4BNR0 A0A2M4BNR5 A0A2M4AHR6 A0A2M4AHW7 N6TKY0 T1HS92 A0A0P4VLB5 A0A182K3D9 W5JQ94 A0A224XRE7 A0A0K8TMY9 A0A182Y414 A0A0V0G7A8 A0A069DT22 A0A0L0CIM7 R4FKT1 A0A1A9UQB6 A0A1B0GA09 A0A0K8V3K1 A0A182QXK9 A0A1A9X8S6 A0A1B0C3A9 A0A034WLM3 A0A0A1XR71 A0A1A9ZEH7 W8CDN6 B4PXA5 A0A1I8NH60 A0A1I8NH52 Q9VXK8 A0A1I8PSJ5 A0A1I8PSG3 B4JX37 A0A1J1J6D2 B3NXG3 A0A336KEK8 A0A182VZV3 B4L6Y3 B4R5K6 A0A1W4VG58 A0A067QQG7 A0A0M4EN53 A0A0A9X8A3 B4MJ77 B3MPW7 A0A1B6DH59 A0A0P9AXA5 B4MA55 A0A0P8YKV4 A0A2R7WJ47 B4HCQ9 D6W807 A0A3B0K191 A0A087ZU62 A0A182GYW9 A0A1B6I2G2 E0VRK5 E2BNR4 A0A0N0BHY1 A0A1Y1LJK4 B0W6T3 A0A084VB74 A0A182FPH8 A0A2A3EH13 A0A1J1J949 E2AE71 A0A151I9T7 A0A026WGK3 A0A151HYQ5 A0A1W4WQ62 F4X481 A0A158N9I2
EC Number
6.1.1.6
Pubmed
22118469
23622113
26354079
29403074
17510324
25474469
+ More
20966253 12364791 14747013 17210077 23537049 27129103 20920257 23761445 26369729 25244985 26334808 26108605 25348373 25830018 24495485 17994087 17550304 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 25401762 26823975 18362917 19820115 26483478 20566863 20798317 28004739 24438588 24508170 30249741 21719571 21347285
20966253 12364791 14747013 17210077 23537049 27129103 20920257 23761445 26369729 25244985 26334808 26108605 25348373 25830018 24495485 17994087 17550304 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 25401762 26823975 18362917 19820115 26483478 20566863 20798317 28004739 24438588 24508170 30249741 21719571 21347285
EMBL
RSAL01000033
RVE51513.1
AGBW02007840
OWR54490.1
GAIX01000275
JAA92285.1
+ More
ODYU01007046 SOQ49472.1 NWSH01001068 PCG72816.1 KQ459585 KPI98373.1 KQ459896 KPJ19364.1 PYGN01000109 PSN54025.1 LJIG01000806 KRT86134.1 CH477529 EAT39488.1 GFDL01001048 JAV33997.1 APCN01000254 GBBI01000993 JAC17719.1 AAAB01008859 EAA08135.6 GANO01003568 JAB56303.1 GGFJ01005816 MBW54957.1 AXCM01000657 GGFK01007121 MBW40442.1 GGFM01003917 MBW24668.1 GGFJ01005578 MBW54719.1 GGFJ01005579 MBW54720.1 GGFK01006999 MBW40320.1 GGFK01007062 MBW40383.1 APGK01020335 KB740193 KB631669 ENN81079.1 ERL85193.1 ACPB03019084 GDKW01003182 JAI53413.1 ADMH02000776 ETN65075.1 GFTR01005845 JAW10581.1 GDAI01002313 JAI15290.1 GECL01002200 JAP03924.1 GBGD01001641 JAC87248.1 JRES01000325 KNC32263.1 GAHY01002364 JAA75146.1 CCAG010018793 GDHF01018835 JAI33479.1 AXCN02001472 JXJN01024889 GAKP01003393 JAC55559.1 GBXI01001214 JAD13078.1 GAMC01001631 JAC04925.1 CM000162 EDX01868.1 AE014298 AY051504 AAF48551.1 AAK92928.1 CH916376 EDV95313.1 CVRI01000072 CRL07530.1 CH954180 EDV46922.1 UFQS01000372 UFQT01000372 SSX03274.1 SSX23640.1 CH933812 EDW06129.1 CM000366 EDX18051.1 KK853048 KDR12085.1 CP012528 ALC49346.1 GBHO01030285 GBHO01030284 GDHC01019906 GDHC01012543 JAG13319.1 JAG13320.1 JAP98722.1 JAQ06086.1 CH963738 EDW72166.2 CH902621 EDV44729.1 GEDC01012281 JAS25017.1 KPU81721.1 CH940655 EDW66114.2 KPU81719.1 KK854868 PTY19351.1 CH479397 EDW29846.1 KQ971307 EFA11016.1 OUUW01000011 SPP86442.1 JXUM01098402 KQ564417 KXJ72322.1 GECU01026585 JAS81121.1 DS235478 EEB16011.1 GL449444 EFN82673.1 KQ435737 KOX76892.1 GEZM01055671 JAV72988.1 DS231849 EDS36914.1 ATLV01006052 KE524335 KFB35218.1 KZ288260 PBC30446.1 CRL07529.1 GL438827 EFN68304.1 KQ978264 KYM95839.1 KK107260 QOIP01000001 EZA54199.1 RLU27262.1 KQ976715 KQ976461 KYM76928.1 KYM84711.1 GL888628 EGI58749.1 ADTU01009655
ODYU01007046 SOQ49472.1 NWSH01001068 PCG72816.1 KQ459585 KPI98373.1 KQ459896 KPJ19364.1 PYGN01000109 PSN54025.1 LJIG01000806 KRT86134.1 CH477529 EAT39488.1 GFDL01001048 JAV33997.1 APCN01000254 GBBI01000993 JAC17719.1 AAAB01008859 EAA08135.6 GANO01003568 JAB56303.1 GGFJ01005816 MBW54957.1 AXCM01000657 GGFK01007121 MBW40442.1 GGFM01003917 MBW24668.1 GGFJ01005578 MBW54719.1 GGFJ01005579 MBW54720.1 GGFK01006999 MBW40320.1 GGFK01007062 MBW40383.1 APGK01020335 KB740193 KB631669 ENN81079.1 ERL85193.1 ACPB03019084 GDKW01003182 JAI53413.1 ADMH02000776 ETN65075.1 GFTR01005845 JAW10581.1 GDAI01002313 JAI15290.1 GECL01002200 JAP03924.1 GBGD01001641 JAC87248.1 JRES01000325 KNC32263.1 GAHY01002364 JAA75146.1 CCAG010018793 GDHF01018835 JAI33479.1 AXCN02001472 JXJN01024889 GAKP01003393 JAC55559.1 GBXI01001214 JAD13078.1 GAMC01001631 JAC04925.1 CM000162 EDX01868.1 AE014298 AY051504 AAF48551.1 AAK92928.1 CH916376 EDV95313.1 CVRI01000072 CRL07530.1 CH954180 EDV46922.1 UFQS01000372 UFQT01000372 SSX03274.1 SSX23640.1 CH933812 EDW06129.1 CM000366 EDX18051.1 KK853048 KDR12085.1 CP012528 ALC49346.1 GBHO01030285 GBHO01030284 GDHC01019906 GDHC01012543 JAG13319.1 JAG13320.1 JAP98722.1 JAQ06086.1 CH963738 EDW72166.2 CH902621 EDV44729.1 GEDC01012281 JAS25017.1 KPU81721.1 CH940655 EDW66114.2 KPU81719.1 KK854868 PTY19351.1 CH479397 EDW29846.1 KQ971307 EFA11016.1 OUUW01000011 SPP86442.1 JXUM01098402 KQ564417 KXJ72322.1 GECU01026585 JAS81121.1 DS235478 EEB16011.1 GL449444 EFN82673.1 KQ435737 KOX76892.1 GEZM01055671 JAV72988.1 DS231849 EDS36914.1 ATLV01006052 KE524335 KFB35218.1 KZ288260 PBC30446.1 CRL07529.1 GL438827 EFN68304.1 KQ978264 KYM95839.1 KK107260 QOIP01000001 EZA54199.1 RLU27262.1 KQ976715 KQ976461 KYM76928.1 KYM84711.1 GL888628 EGI58749.1 ADTU01009655
Proteomes
UP000283053
UP000007151
UP000218220
UP000053268
UP000053240
UP000245037
+ More
UP000008820 UP000075840 UP000075903 UP000075882 UP000007062 UP000076407 UP000075902 UP000075900 UP000075885 UP000075883 UP000075884 UP000019118 UP000030742 UP000015103 UP000075881 UP000000673 UP000076408 UP000037069 UP000078200 UP000092444 UP000075886 UP000092443 UP000092460 UP000092445 UP000002282 UP000095301 UP000000803 UP000095300 UP000001070 UP000183832 UP000008711 UP000075920 UP000009192 UP000000304 UP000192221 UP000027135 UP000092553 UP000007798 UP000007801 UP000008792 UP000008744 UP000007266 UP000268350 UP000005203 UP000069940 UP000249989 UP000009046 UP000008237 UP000053105 UP000002320 UP000030765 UP000069272 UP000242457 UP000000311 UP000078542 UP000053097 UP000279307 UP000078540 UP000192223 UP000007755 UP000005205
UP000008820 UP000075840 UP000075903 UP000075882 UP000007062 UP000076407 UP000075902 UP000075900 UP000075885 UP000075883 UP000075884 UP000019118 UP000030742 UP000015103 UP000075881 UP000000673 UP000076408 UP000037069 UP000078200 UP000092444 UP000075886 UP000092443 UP000092460 UP000092445 UP000002282 UP000095301 UP000000803 UP000095300 UP000001070 UP000183832 UP000008711 UP000075920 UP000009192 UP000000304 UP000192221 UP000027135 UP000092553 UP000007798 UP000007801 UP000008792 UP000008744 UP000007266 UP000268350 UP000005203 UP000069940 UP000249989 UP000009046 UP000008237 UP000053105 UP000002320 UP000030765 UP000069272 UP000242457 UP000000311 UP000078542 UP000053097 UP000279307 UP000078540 UP000192223 UP000007755 UP000005205
PRIDE
Pfam
Interpro
IPR018108
Mitochondrial_sb/sol_carrier
+ More
IPR023395 Mt_carrier_dom_sf
IPR039158 SLC25A46
IPR005033 YEATS
IPR038704 YEAST_sf
IPR040930 AF-9_AHD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR012934 Znf_AD
IPR004365 NA-bd_OB_tRNA
IPR004364 aa-tRNA-synt_II
IPR018149 Lys-tRNA-synth_II_C
IPR012340 NA-bd_OB-fold
IPR006195 aa-tRNA-synth_II
IPR002313 Lys-tRNA-ligase_II
IPR023395 Mt_carrier_dom_sf
IPR039158 SLC25A46
IPR005033 YEATS
IPR038704 YEAST_sf
IPR040930 AF-9_AHD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR012934 Znf_AD
IPR004365 NA-bd_OB_tRNA
IPR004364 aa-tRNA-synt_II
IPR018149 Lys-tRNA-synth_II_C
IPR012340 NA-bd_OB-fold
IPR006195 aa-tRNA-synth_II
IPR002313 Lys-tRNA-ligase_II
Gene 3D
ProteinModelPortal
A0A3S2PHC9
A0A212FLC2
S4PS34
A0A2H1W9F0
A0A2A4JM08
A0A194PZA2
+ More
A0A194RPN1 A0A2P8ZC01 A0A0T6BFM2 Q16XZ0 A0A1S4FK72 A0A1Q3G2F2 A0A182HS89 A0A023F9P1 A0A182V973 A0A182LE70 Q7PQY0 U5ERT7 A0A182XAF0 A0A182U9G6 A0A182RTI5 A0A182PL56 A0A2M4BPF6 A0A182LV47 A0A182NR60 A0A2M4AI87 A0A2M3Z8A0 A0A2M4BNR0 A0A2M4BNR5 A0A2M4AHR6 A0A2M4AHW7 N6TKY0 T1HS92 A0A0P4VLB5 A0A182K3D9 W5JQ94 A0A224XRE7 A0A0K8TMY9 A0A182Y414 A0A0V0G7A8 A0A069DT22 A0A0L0CIM7 R4FKT1 A0A1A9UQB6 A0A1B0GA09 A0A0K8V3K1 A0A182QXK9 A0A1A9X8S6 A0A1B0C3A9 A0A034WLM3 A0A0A1XR71 A0A1A9ZEH7 W8CDN6 B4PXA5 A0A1I8NH60 A0A1I8NH52 Q9VXK8 A0A1I8PSJ5 A0A1I8PSG3 B4JX37 A0A1J1J6D2 B3NXG3 A0A336KEK8 A0A182VZV3 B4L6Y3 B4R5K6 A0A1W4VG58 A0A067QQG7 A0A0M4EN53 A0A0A9X8A3 B4MJ77 B3MPW7 A0A1B6DH59 A0A0P9AXA5 B4MA55 A0A0P8YKV4 A0A2R7WJ47 B4HCQ9 D6W807 A0A3B0K191 A0A087ZU62 A0A182GYW9 A0A1B6I2G2 E0VRK5 E2BNR4 A0A0N0BHY1 A0A1Y1LJK4 B0W6T3 A0A084VB74 A0A182FPH8 A0A2A3EH13 A0A1J1J949 E2AE71 A0A151I9T7 A0A026WGK3 A0A151HYQ5 A0A1W4WQ62 F4X481 A0A158N9I2
A0A194RPN1 A0A2P8ZC01 A0A0T6BFM2 Q16XZ0 A0A1S4FK72 A0A1Q3G2F2 A0A182HS89 A0A023F9P1 A0A182V973 A0A182LE70 Q7PQY0 U5ERT7 A0A182XAF0 A0A182U9G6 A0A182RTI5 A0A182PL56 A0A2M4BPF6 A0A182LV47 A0A182NR60 A0A2M4AI87 A0A2M3Z8A0 A0A2M4BNR0 A0A2M4BNR5 A0A2M4AHR6 A0A2M4AHW7 N6TKY0 T1HS92 A0A0P4VLB5 A0A182K3D9 W5JQ94 A0A224XRE7 A0A0K8TMY9 A0A182Y414 A0A0V0G7A8 A0A069DT22 A0A0L0CIM7 R4FKT1 A0A1A9UQB6 A0A1B0GA09 A0A0K8V3K1 A0A182QXK9 A0A1A9X8S6 A0A1B0C3A9 A0A034WLM3 A0A0A1XR71 A0A1A9ZEH7 W8CDN6 B4PXA5 A0A1I8NH60 A0A1I8NH52 Q9VXK8 A0A1I8PSJ5 A0A1I8PSG3 B4JX37 A0A1J1J6D2 B3NXG3 A0A336KEK8 A0A182VZV3 B4L6Y3 B4R5K6 A0A1W4VG58 A0A067QQG7 A0A0M4EN53 A0A0A9X8A3 B4MJ77 B3MPW7 A0A1B6DH59 A0A0P9AXA5 B4MA55 A0A0P8YKV4 A0A2R7WJ47 B4HCQ9 D6W807 A0A3B0K191 A0A087ZU62 A0A182GYW9 A0A1B6I2G2 E0VRK5 E2BNR4 A0A0N0BHY1 A0A1Y1LJK4 B0W6T3 A0A084VB74 A0A182FPH8 A0A2A3EH13 A0A1J1J949 E2AE71 A0A151I9T7 A0A026WGK3 A0A151HYQ5 A0A1W4WQ62 F4X481 A0A158N9I2
PDB
4C9Q
E-value=0.0160614,
Score=90
Ontologies
GO
Topology
Subcellular location
Nucleus
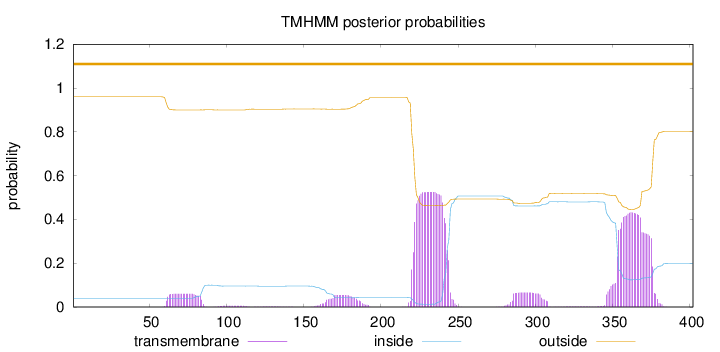
Length:
402
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
25.34436
Exp number, first 60 AAs:
0.00471
Total prob of N-in:
0.04020
outside
1 - 402
Population Genetic Test Statistics
Pi
50.254339
Theta
153.502769
Tajima's D
-0.431333
CLR
14.47765
CSRT
0.259737013149343
Interpretation
Uncertain
