Gene
KWMTBOMO12466
Pre Gene Modal
BGIBMGA001493
Annotation
PREDICTED:_voltage-dependent_calcium_channel_subunit_alpha-2/delta-3-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.408 Nuclear Reliability : 1.947
Sequence
CDS
ATGAAAATTAAGCTAATCGTACCGATTATACTAACAGCTATACTCATTCGATCAACTGAATCTGTAAAACATAAAAATGTACTTCAAAATAGAACTGTAGCCGAAGTGAAAAATAGTCTCGAGAAACACGAGGACGCTTTGTCCGAAGTGCTCAAGCACAAAACCGGAGTTAACCCGATAAAAATTACTAATGTAACTCAAGCGAAAATTAAGCCTTTCGTCAAGAAAGAAAAAAAGATTCTAAGAATGATTTCAACGAAATCGATCGAAAATTGGGCACAAAGGCTCTCCGAAGTATTCTTCGAGATCGAAAGCAAGCTTGTTAAGCGAGAGGAACTATTGAGCGCATTTAGCGATGCTATGATGGTAGCTCGCAACGGAACCGCCCTCGTGAACAAGGCTGCAGACGCCTTAGAAGACTTGTTAACAAGAAGAGTCAAAGCCGCCGAAAATATAATGCGCAAAGCTGAAGAATTAGATAAGGACGAAACACCTCCGCCTGACAATTACGTTTTTGACCATAGCGTGGATTTAGATAAAATCAAGGAGAAGAAGATACCCAATCAGAAAGGCTGGTTCATGCCACAGAACTGTACTAATTTACAAAAGGTACCCTTGAACGAGAGTAATCATTTCGATGGCAAAGTATCACTCCAGACGTCCAGCGTTTATGTTTCGCTTGAAGTTTTTGAATGCGATCCTAAGGTTATGAAACATATATATTGGACGGAAGGGTTATTGTCGACATTCAAAAAGAATTACGCGCAGGACGCGACCACGGACTTTCAGTATTTCTGTAGTGCTAAAGGATTCTTACGACACTACCCAGCTGCTTTATGGGACGACATGCACCAGCTCAAGTTGAATACGGACGACGTGTACGACTGTCGGCTGCGGTCGTGGTACGTGAGCGCCGGCGGAGCGCCCCGTGACGTACTCGTGTTGCTGGACGCCTCGGGCTCCATGAACGGCTCCTCGAACCAGGTCATCGCTGAGGAGTTCACTTTGGCTCTGCTCAGCGCTCTCACCGACGACGATCAAGTGAACGTCTTATACTTCAATGAGAAAGTTAATTCGCTTATCCAGTGCTTCAATCAGAAAATGGTTCCGGCAAACCACGTGAATTCTGCCGCGATGATGGAAGAGCTAAGGGAATACACGAAAGTAAACGAGACGAAAGTGGACTACGCGCTAAAGTACGCGATAAAACTGCTACGGAAACAGCGCTTGATTAGAGACCGACCGAAGTCGTGTCAGCAAGCTATAGTGCTGCTCACGGACAGCATGTACGACAATTACACGGACCTCGTGAGGCATTTGGACCCCGGGAATAATATCAGACTGTTCGTGTTCTGGCTGCACGACCGGTACGGGCTGCGCGACGACACGCGGCGGCTCGCGGAGTGGCTGAGCTGCGCGCGCGACGGCTACTTCGCCGAGCTCATCACGCACGCTGACGTCACGGAGCAGGTCATGAGCGTGCTGCGCGTGCTGGTTCGCCCGCTCGTCGCGCAGCGGGACCACAGGCTGCGGGTCTTCAGCGATATATACGCTCACATTGAGGACCCGAGACGCAGCGAGTACCACTGGACGCAGAAGGAGAACGCGGAACAACTCTATCGTTACAAGGAGCTGAGGAAGGACAAACGCAAACTCCTCGACCCGAGGCGAATGTATGAAGATTGGATGCACCAATGGAAACTCAACGAATACGGGCAGTATTATGAAGGAGAAGAATTGAATTATCGTTTGGAAATAAGCGTTTCTGTGCCCGTTTTTGATGCTACTACGGCAGAGAATATTACGATCCAATTGATGGAGGAGAGACAACGGAACTCTACGAGACAGTATCCAGTAAACCGTCTTCTGGGAGTAGCAGGAGTGGACATTCCAATCGACCATTTGAAGTTGATTCTGCCGTATCATCAGCTAGGTGCGGGCGGCTCAATCATACTGGTGGATCATCGTGGAAACGTTGTCTTGCACGACAATTTGAAACCTACATTCGACGGAGACGTCTTGAGGCCTGGATACAGGACCGTTGACTTCCTCGACTTGGAACAGCCGGCAGTCAGCCACCTCCCCAGACAATACTTGCCAGATTGGCTCAAGTTTCGACGAAATCTGATAATCGATAAAGAAACTGGCAACATGACCATGTACGCGAAAAACATTTTCGAAGACGGAATGAGGGCAACGTTGGAAAATCGGCAATACTTTTGGAGAAGGGTCCTGGACCACTACACCGTGGTCGTCGCTCTACCGAACCCCGACGCGCACTGGGCCGGGTCCGAGTCCCAGTTCACCATGAAGCTCGCTGAAGCGGCGTTGAAGTCGCTGTCCCGAACCGACTTCGCCGTGCAGCCAGACTGGCTGTACTGCCGGCACGTGGAGCCCCACTTCGACTCCCGCGAGGCCGAGGTGCTCCACTTCATCAGGCGGCGGAGCGACGAGCCCAACTTCGCGATGAAGAAGCTGAATCATATATTCTCACCCATCCCACCTTTGTTACTCGAAAAAACATATCAATGTGATGAAGGTCTAATGGCGAGGCTGTGCAAGGACGCCATAGCCACGGACAAGTGGGCCCGGGAGCACGAGCACCCGCACCGGGCCCGGGACTGCTCCACCTGCGAGCTGGGCTCCATCACCACCTTCTTCGCGAGTGAGAACGGGCTGACCCGATGGCAGGTGCACCACGCGACCAGCGAGCACGCGGCGCCCGTGGAGGGCAGCGTGTGGGCGCGCGGGCCGCGGGAGGCGTGGTACCGGCGCGCCGCGCACCGCCCCGCCGCGCTGCACGTGCACGCGCCCGCGCGCCGCCCGCAGCTGCGCCGGAACAGCTTCGCGCCGCCCCCGCCGGTGCCTGTCAGCCAACAATGGCTAACGGCTGCTCGAGCCCTCACCTCCCCGGACAAGGGCATCATCGGTGTAACCGGCTTCCACTTCTATCCGAGCCACCTGGACGATTTACTCAACAGCATCACCAAATTTCCGGTGAGTTTTAGCTAA
Protein
MKIKLIVPIILTAILIRSTESVKHKNVLQNRTVAEVKNSLEKHEDALSEVLKHKTGVNPIKITNVTQAKIKPFVKKEKKILRMISTKSIENWAQRLSEVFFEIESKLVKREELLSAFSDAMMVARNGTALVNKAADALEDLLTRRVKAAENIMRKAEELDKDETPPPDNYVFDHSVDLDKIKEKKIPNQKGWFMPQNCTNLQKVPLNESNHFDGKVSLQTSSVYVSLEVFECDPKVMKHIYWTEGLLSTFKKNYAQDATTDFQYFCSAKGFLRHYPAALWDDMHQLKLNTDDVYDCRLRSWYVSAGGAPRDVLVLLDASGSMNGSSNQVIAEEFTLALLSALTDDDQVNVLYFNEKVNSLIQCFNQKMVPANHVNSAAMMEELREYTKVNETKVDYALKYAIKLLRKQRLIRDRPKSCQQAIVLLTDSMYDNYTDLVRHLDPGNNIRLFVFWLHDRYGLRDDTRRLAEWLSCARDGYFAELITHADVTEQVMSVLRVLVRPLVAQRDHRLRVFSDIYAHIEDPRRSEYHWTQKENAEQLYRYKELRKDKRKLLDPRRMYEDWMHQWKLNEYGQYYEGEELNYRLEISVSVPVFDATTAENITIQLMEERQRNSTRQYPVNRLLGVAGVDIPIDHLKLILPYHQLGAGGSIILVDHRGNVVLHDNLKPTFDGDVLRPGYRTVDFLDLEQPAVSHLPRQYLPDWLKFRRNLIIDKETGNMTMYAKNIFEDGMRATLENRQYFWRRVLDHYTVVVALPNPDAHWAGSESQFTMKLAEAALKSLSRTDFAVQPDWLYCRHVEPHFDSREAEVLHFIRRRSDEPNFAMKKLNHIFSPIPPLLLEKTYQCDEGLMARLCKDAIATDKWAREHEHPHRARDCSTCELGSITTFFASENGLTRWQVHHATSEHAAPVEGSVWARGPREAWYRRAAHRPAALHVHAPARRPQLRRNSFAPPPPVPVSQQWLTAARALTSPDKGIIGVTGFHFYPSHLDDLLNSITKFPVSFS
Summary
Uniprot
H9IW62
A0A2H1W8S9
A0A2A4JQG6
A0A437BM10
A0A212EJI2
A0A194RTC6
+ More
A0A194Q048 A0A3S2M507 A0A212F2P3 A0A2A4JW43 A0A2A4JVU0 A0A0N1IFN0 A0A2H1W0S0 A0A151J7L7 F4WDF0 A0A3L8DHW5 A0A088A0C5 A0A0L7R1P7 A0A026WML2 A0A0B5EIM5 A0A1S4EV02 Q7Q758 B0WES9 W5J6G9 A0A182QGK0 A0A182WKA1 A0A182Y0W5 A0A2W1BPP2 A0A182LJL0 A0A194PP00 A0A182UNW2 A0A182J7M5 W8BYH9 A0A1S4GQ08 A0A195FRY6 H9J2A3 A0A2A3EQI3 A0A154P370 Q17Q76 E2BCL9 A0A084W2X5 A0A034VAZ5 A0A0R1DPV7 A0A336LKR3 E2AL65 A0A2J7PJ67 A0A0P8Y6U5 A0A0Q5VUW3 A0A0B4K7P4 A0A1B0FGH7 A0A0P8ZRP9 A0A1J1HY10 A0A0R1DWW0 A0A1W4XCA0 A0A1W4VHY6 A0A0Q5VNY3 A0A034VFB1 A0A1W4VH47 A0A336LKI8 A0A336LHN4 B4P469 A0A2J7PJ48 A0A0Q9X2P9 A0A0P8YDV8 A0A1W4VH43 A0A1W4VGK1 A0A0Q5VMU8 A0A1I8QDT3 A0A067RGR1 A0A0Q5VMW2 Q7K0H4 N6WCF7 A0A195CBL9 B3MHE1 A0A0J9RD72 A0A0B4K866 A0A0Q9W3C2 A0A182UDJ5 A0A0Q9W3E8 B3NRH4 A0A1I8QDU6 A0A0L0CQA0 A0A0P9C0N1 A0A1I8QDV5 B4N5V0 A0A1I8N322 A0A1W4V4S8 A0A1W4V4S3 K7IUL2 A0A0Q9X480 A0A1W4V4B9 A0A182RN46 A0A0Q9WF22 Q28XX9 B4LKL2 A0A0J9RC37 A0A0M8ZSY5 A0A182XIL0 A0A1I8QDU9 A0A0Q9WEY5
A0A194Q048 A0A3S2M507 A0A212F2P3 A0A2A4JW43 A0A2A4JVU0 A0A0N1IFN0 A0A2H1W0S0 A0A151J7L7 F4WDF0 A0A3L8DHW5 A0A088A0C5 A0A0L7R1P7 A0A026WML2 A0A0B5EIM5 A0A1S4EV02 Q7Q758 B0WES9 W5J6G9 A0A182QGK0 A0A182WKA1 A0A182Y0W5 A0A2W1BPP2 A0A182LJL0 A0A194PP00 A0A182UNW2 A0A182J7M5 W8BYH9 A0A1S4GQ08 A0A195FRY6 H9J2A3 A0A2A3EQI3 A0A154P370 Q17Q76 E2BCL9 A0A084W2X5 A0A034VAZ5 A0A0R1DPV7 A0A336LKR3 E2AL65 A0A2J7PJ67 A0A0P8Y6U5 A0A0Q5VUW3 A0A0B4K7P4 A0A1B0FGH7 A0A0P8ZRP9 A0A1J1HY10 A0A0R1DWW0 A0A1W4XCA0 A0A1W4VHY6 A0A0Q5VNY3 A0A034VFB1 A0A1W4VH47 A0A336LKI8 A0A336LHN4 B4P469 A0A2J7PJ48 A0A0Q9X2P9 A0A0P8YDV8 A0A1W4VH43 A0A1W4VGK1 A0A0Q5VMU8 A0A1I8QDT3 A0A067RGR1 A0A0Q5VMW2 Q7K0H4 N6WCF7 A0A195CBL9 B3MHE1 A0A0J9RD72 A0A0B4K866 A0A0Q9W3C2 A0A182UDJ5 A0A0Q9W3E8 B3NRH4 A0A1I8QDU6 A0A0L0CQA0 A0A0P9C0N1 A0A1I8QDV5 B4N5V0 A0A1I8N322 A0A1W4V4S8 A0A1W4V4S3 K7IUL2 A0A0Q9X480 A0A1W4V4B9 A0A182RN46 A0A0Q9WF22 Q28XX9 B4LKL2 A0A0J9RC37 A0A0M8ZSY5 A0A182XIL0 A0A1I8QDU9 A0A0Q9WEY5
Pubmed
19121390
22118469
26354079
21719571
30249741
24508170
+ More
17510324 12364791 20920257 23761445 25244985 28756777 20966253 24495485 20798317 24438588 25348373 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24845553 26109357 26109356 15632085 22936249 26108605 25315136 20075255
17510324 12364791 20920257 23761445 25244985 28756777 20966253 24495485 20798317 24438588 25348373 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24845553 26109357 26109356 15632085 22936249 26108605 25315136 20075255
EMBL
BABH01005884
BABH01005885
BABH01005886
BABH01005887
ODYU01007046
SOQ49470.1
+ More
NWSH01000891 PCG73662.1 RSAL01000033 RVE51510.1 AGBW02014451 OWR41649.1 KQ459896 KPJ19361.1 KQ459585 KPI98369.1 RSAL01000038 RVE51090.1 AGBW02010703 OWR48004.1 NWSH01000568 PCG75602.1 PCG75603.1 KQ460226 KPJ16443.1 ODYU01005348 SOQ46104.1 KQ979695 KYN19721.1 GL888087 EGI67730.1 QOIP01000007 RLU20007.1 KQ414667 KOC64769.1 KK107152 EZA57148.1 KJ485707 AJE70859.1 AAAB01008960 EAA10906.4 DS231911 EDS45842.1 ADMH02002072 ETN59596.1 AXCN02000940 KZ149992 PZC75535.1 KQ459597 KPI95037.1 GAMC01012241 JAB94314.1 KQ981285 KYN43191.1 BABH01007564 BABH01007565 KZ288194 PBC33967.1 KQ434809 KZC06283.1 CH477187 EAT48840.1 GL447336 EFN86529.1 ATLV01019720 KE525278 KFB44569.1 GAKP01018466 JAC40486.1 CM000158 KRJ99303.1 UFQT01000004 SSX17249.1 GL440492 EFN65820.1 NEVH01024950 PNF16364.1 CH902619 KPU77120.1 CH954179 KQS62709.1 AE013599 AFH08058.1 CCAG010013633 CCAG010013634 KPU77119.1 CVRI01000036 CRK92984.1 KRJ99302.1 KQS62708.1 GAKP01018467 JAC40485.1 SSX17251.1 SSX17250.1 EDW90579.2 PNF16365.1 CH964154 KRF99110.1 KPU77117.1 KQS62711.1 KK852476 KDR23041.1 KQS62710.1 AY069830 AAF58335.2 AAL39975.1 CM000071 ENO01819.1 KQ978068 KYM97596.1 EDV37941.1 CM002911 KMY93564.1 AFH08057.1 CH940648 KRF79594.1 KRF79596.1 EDV56126.2 JRES01000080 KNC34362.1 KPU77118.1 EDW79739.1 AAZX01002632 KRF99111.1 KRF79598.1 EAL26187.3 EDW60733.1 KMY93563.1 KQ435863 KOX70317.1 KRF79595.1
NWSH01000891 PCG73662.1 RSAL01000033 RVE51510.1 AGBW02014451 OWR41649.1 KQ459896 KPJ19361.1 KQ459585 KPI98369.1 RSAL01000038 RVE51090.1 AGBW02010703 OWR48004.1 NWSH01000568 PCG75602.1 PCG75603.1 KQ460226 KPJ16443.1 ODYU01005348 SOQ46104.1 KQ979695 KYN19721.1 GL888087 EGI67730.1 QOIP01000007 RLU20007.1 KQ414667 KOC64769.1 KK107152 EZA57148.1 KJ485707 AJE70859.1 AAAB01008960 EAA10906.4 DS231911 EDS45842.1 ADMH02002072 ETN59596.1 AXCN02000940 KZ149992 PZC75535.1 KQ459597 KPI95037.1 GAMC01012241 JAB94314.1 KQ981285 KYN43191.1 BABH01007564 BABH01007565 KZ288194 PBC33967.1 KQ434809 KZC06283.1 CH477187 EAT48840.1 GL447336 EFN86529.1 ATLV01019720 KE525278 KFB44569.1 GAKP01018466 JAC40486.1 CM000158 KRJ99303.1 UFQT01000004 SSX17249.1 GL440492 EFN65820.1 NEVH01024950 PNF16364.1 CH902619 KPU77120.1 CH954179 KQS62709.1 AE013599 AFH08058.1 CCAG010013633 CCAG010013634 KPU77119.1 CVRI01000036 CRK92984.1 KRJ99302.1 KQS62708.1 GAKP01018467 JAC40485.1 SSX17251.1 SSX17250.1 EDW90579.2 PNF16365.1 CH964154 KRF99110.1 KPU77117.1 KQS62711.1 KK852476 KDR23041.1 KQS62710.1 AY069830 AAF58335.2 AAL39975.1 CM000071 ENO01819.1 KQ978068 KYM97596.1 EDV37941.1 CM002911 KMY93564.1 AFH08057.1 CH940648 KRF79594.1 KRF79596.1 EDV56126.2 JRES01000080 KNC34362.1 KPU77118.1 EDW79739.1 AAZX01002632 KRF99111.1 KRF79598.1 EAL26187.3 EDW60733.1 KMY93563.1 KQ435863 KOX70317.1 KRF79595.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000078492 UP000007755 UP000279307 UP000005203 UP000053825 UP000053097 UP000007062 UP000002320 UP000000673 UP000075886 UP000075920 UP000076408 UP000075882 UP000075903 UP000075880 UP000078541 UP000242457 UP000076502 UP000008820 UP000008237 UP000030765 UP000002282 UP000000311 UP000235965 UP000007801 UP000008711 UP000000803 UP000092444 UP000183832 UP000192223 UP000192221 UP000007798 UP000095300 UP000027135 UP000001819 UP000078542 UP000008792 UP000075902 UP000037069 UP000095301 UP000002358 UP000075900 UP000053105 UP000076407
UP000078492 UP000007755 UP000279307 UP000005203 UP000053825 UP000053097 UP000007062 UP000002320 UP000000673 UP000075886 UP000075920 UP000076408 UP000075882 UP000075903 UP000075880 UP000078541 UP000242457 UP000076502 UP000008820 UP000008237 UP000030765 UP000002282 UP000000311 UP000235965 UP000007801 UP000008711 UP000000803 UP000092444 UP000183832 UP000192223 UP000192221 UP000007798 UP000095300 UP000027135 UP000001819 UP000078542 UP000008792 UP000075902 UP000037069 UP000095301 UP000002358 UP000075900 UP000053105 UP000076407
Pfam
Interpro
SUPFAM
SSF53300
SSF53300
Gene 3D
ProteinModelPortal
H9IW62
A0A2H1W8S9
A0A2A4JQG6
A0A437BM10
A0A212EJI2
A0A194RTC6
+ More
A0A194Q048 A0A3S2M507 A0A212F2P3 A0A2A4JW43 A0A2A4JVU0 A0A0N1IFN0 A0A2H1W0S0 A0A151J7L7 F4WDF0 A0A3L8DHW5 A0A088A0C5 A0A0L7R1P7 A0A026WML2 A0A0B5EIM5 A0A1S4EV02 Q7Q758 B0WES9 W5J6G9 A0A182QGK0 A0A182WKA1 A0A182Y0W5 A0A2W1BPP2 A0A182LJL0 A0A194PP00 A0A182UNW2 A0A182J7M5 W8BYH9 A0A1S4GQ08 A0A195FRY6 H9J2A3 A0A2A3EQI3 A0A154P370 Q17Q76 E2BCL9 A0A084W2X5 A0A034VAZ5 A0A0R1DPV7 A0A336LKR3 E2AL65 A0A2J7PJ67 A0A0P8Y6U5 A0A0Q5VUW3 A0A0B4K7P4 A0A1B0FGH7 A0A0P8ZRP9 A0A1J1HY10 A0A0R1DWW0 A0A1W4XCA0 A0A1W4VHY6 A0A0Q5VNY3 A0A034VFB1 A0A1W4VH47 A0A336LKI8 A0A336LHN4 B4P469 A0A2J7PJ48 A0A0Q9X2P9 A0A0P8YDV8 A0A1W4VH43 A0A1W4VGK1 A0A0Q5VMU8 A0A1I8QDT3 A0A067RGR1 A0A0Q5VMW2 Q7K0H4 N6WCF7 A0A195CBL9 B3MHE1 A0A0J9RD72 A0A0B4K866 A0A0Q9W3C2 A0A182UDJ5 A0A0Q9W3E8 B3NRH4 A0A1I8QDU6 A0A0L0CQA0 A0A0P9C0N1 A0A1I8QDV5 B4N5V0 A0A1I8N322 A0A1W4V4S8 A0A1W4V4S3 K7IUL2 A0A0Q9X480 A0A1W4V4B9 A0A182RN46 A0A0Q9WF22 Q28XX9 B4LKL2 A0A0J9RC37 A0A0M8ZSY5 A0A182XIL0 A0A1I8QDU9 A0A0Q9WEY5
A0A194Q048 A0A3S2M507 A0A212F2P3 A0A2A4JW43 A0A2A4JVU0 A0A0N1IFN0 A0A2H1W0S0 A0A151J7L7 F4WDF0 A0A3L8DHW5 A0A088A0C5 A0A0L7R1P7 A0A026WML2 A0A0B5EIM5 A0A1S4EV02 Q7Q758 B0WES9 W5J6G9 A0A182QGK0 A0A182WKA1 A0A182Y0W5 A0A2W1BPP2 A0A182LJL0 A0A194PP00 A0A182UNW2 A0A182J7M5 W8BYH9 A0A1S4GQ08 A0A195FRY6 H9J2A3 A0A2A3EQI3 A0A154P370 Q17Q76 E2BCL9 A0A084W2X5 A0A034VAZ5 A0A0R1DPV7 A0A336LKR3 E2AL65 A0A2J7PJ67 A0A0P8Y6U5 A0A0Q5VUW3 A0A0B4K7P4 A0A1B0FGH7 A0A0P8ZRP9 A0A1J1HY10 A0A0R1DWW0 A0A1W4XCA0 A0A1W4VHY6 A0A0Q5VNY3 A0A034VFB1 A0A1W4VH47 A0A336LKI8 A0A336LHN4 B4P469 A0A2J7PJ48 A0A0Q9X2P9 A0A0P8YDV8 A0A1W4VH43 A0A1W4VGK1 A0A0Q5VMU8 A0A1I8QDT3 A0A067RGR1 A0A0Q5VMW2 Q7K0H4 N6WCF7 A0A195CBL9 B3MHE1 A0A0J9RD72 A0A0B4K866 A0A0Q9W3C2 A0A182UDJ5 A0A0Q9W3E8 B3NRH4 A0A1I8QDU6 A0A0L0CQA0 A0A0P9C0N1 A0A1I8QDV5 B4N5V0 A0A1I8N322 A0A1W4V4S8 A0A1W4V4S3 K7IUL2 A0A0Q9X480 A0A1W4V4B9 A0A182RN46 A0A0Q9WF22 Q28XX9 B4LKL2 A0A0J9RC37 A0A0M8ZSY5 A0A182XIL0 A0A1I8QDU9 A0A0Q9WEY5
PDB
5GJW
E-value=1.13021e-39,
Score=414
Ontologies
GO
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.918213
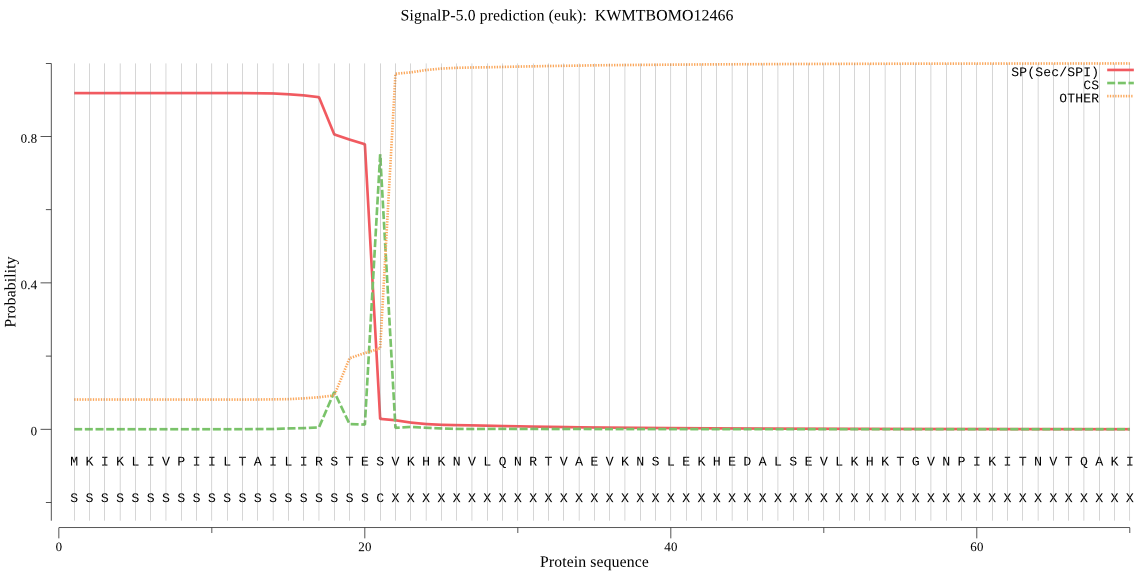
Length:
1003
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0423000000000001
Exp number, first 60 AAs:
0.04204
Total prob of N-in:
0.00246
outside
1 - 1003
Population Genetic Test Statistics
Pi
1.526499
Theta
226.210547
Tajima's D
-2.334395
CLR
220.517958
CSRT
0.00259987000649968
Interpretation
Possibly Positive selection
