Gene
KWMTBOMO12460 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001483
Annotation
PREDICTED:_40S_ribosomal_protein_S17_[Amyelois_transitella]
Full name
40S ribosomal protein S17
Location in the cell
Nuclear Reliability : 2.455
Sequence
CDS
ATGTGTGTAAAAAGATTTGCCACACATTTAATGAGGCGTCTCAGACACTCGCAAGTGCGAGGAATCTCTATCAAACTTCAGGAAGAGGAGCGTGAGAGGCGTGACAACTATGTCCCAGAAGTGTCTGCTCTCGAACATGACATCATCGAAGTAGACCCCGACACCAAGGATATGTTGAAGATGCTGGACTTCAACAATATTAATGGCCTGCAACTAACACAGCCAGCTACCCAGGGTGGCTATGGTGGCAGGCGTAATTAA
Protein
MCVKRFATHLMRRLRHSQVRGISIKLQEEERERRDNYVPEVSALEHDIIEVDPDTKDMLKMLDFNNINGLQLTQPATQGGYGGRRN
Summary
Similarity
Belongs to the eukaryotic ribosomal protein eS17 family.
Keywords
Ribonucleoprotein
Ribosomal protein
Feature
chain 40S ribosomal protein S17
Uniprot
A0A194RGC2
A0A2A4JYV9
I4DMT0
A0A437BMI5
A0A1E1WIM7
I4DIP7
+ More
Q5UAM5 D1LYL3 Q962R2 H9IW52 A0A194Q038 A7KCW4 G0ZEH0 A0A2H1WJA7 A0A212F614 E7DZ51 A0A0L7KZF2 A0A3G1T1N0 Q6F467 A0A067RHS7 A0A336LT32 A0A2P8ZCR8 Q4GXQ2 A0A2J7PYH7 A0A146M2H0 A5HMP2 A0A0A9Z7U0 D1M830 E0V9P6 A0A1D1XQ01 A0A1W4WSI4 Q6EUY4 E9ID71 E2BLN4 A0A195BWS5 E2AYV2 A0A151IEF9 D3KD00 A0A158NEI8 A0A146L5M2 Q6EUY6 Q6EUY5 A0A195FKI1 A0A026W1D6 D6WTW8 A0A1Y1MJZ0 A0A0J7L9E1 A0A3L8DCQ1 F4WRX8 A0A1B6MM82 A0A3L8DGT6 K7JAI6 V5H141 N6SYI9 Q0PXZ4 A0A1B6EUI2 A0A1B6HUL3 A0A1B6CKU2 A0A026WXT7 A0A1Q3FAB7 U5EXA4 B0WC62 J3JYN8 A0A0K8TPU7 U5EZC1 A0A1I8QF05 A0A0C9R7K5 A0A034WH81 A0A0A1WXJ6 A0A0K8V497 A0A0P6GVU4 A0A1L8DRY8 A0A0P4VME1 A0A0V0G4X0 R4G4C3 A6YPF7 A0A1B0D409 A0A0A0R236 B4MMS0 A0A3B0JVC5 Q29FA0 B4H1L6 A0A154NY80 A0A0N0BI35 A0A2A3EHS2 V9IHR8 A0A088ANM8 A0A1I8MSV1 A8C9W2 A0A224XZZ9 A0A1L8EF83 A0A0C9R6X9 A0A0M9A475 A0A1L8EFD0 A0A0L7REE3 A0A3B0J6C8 A0A0X7YAT6 A0A182IZ62 R4WND8 A0A0A0R243
Q5UAM5 D1LYL3 Q962R2 H9IW52 A0A194Q038 A7KCW4 G0ZEH0 A0A2H1WJA7 A0A212F614 E7DZ51 A0A0L7KZF2 A0A3G1T1N0 Q6F467 A0A067RHS7 A0A336LT32 A0A2P8ZCR8 Q4GXQ2 A0A2J7PYH7 A0A146M2H0 A5HMP2 A0A0A9Z7U0 D1M830 E0V9P6 A0A1D1XQ01 A0A1W4WSI4 Q6EUY4 E9ID71 E2BLN4 A0A195BWS5 E2AYV2 A0A151IEF9 D3KD00 A0A158NEI8 A0A146L5M2 Q6EUY6 Q6EUY5 A0A195FKI1 A0A026W1D6 D6WTW8 A0A1Y1MJZ0 A0A0J7L9E1 A0A3L8DCQ1 F4WRX8 A0A1B6MM82 A0A3L8DGT6 K7JAI6 V5H141 N6SYI9 Q0PXZ4 A0A1B6EUI2 A0A1B6HUL3 A0A1B6CKU2 A0A026WXT7 A0A1Q3FAB7 U5EXA4 B0WC62 J3JYN8 A0A0K8TPU7 U5EZC1 A0A1I8QF05 A0A0C9R7K5 A0A034WH81 A0A0A1WXJ6 A0A0K8V497 A0A0P6GVU4 A0A1L8DRY8 A0A0P4VME1 A0A0V0G4X0 R4G4C3 A6YPF7 A0A1B0D409 A0A0A0R236 B4MMS0 A0A3B0JVC5 Q29FA0 B4H1L6 A0A154NY80 A0A0N0BI35 A0A2A3EHS2 V9IHR8 A0A088ANM8 A0A1I8MSV1 A8C9W2 A0A224XZZ9 A0A1L8EF83 A0A0C9R6X9 A0A0M9A475 A0A1L8EFD0 A0A0L7REE3 A0A3B0J6C8 A0A0X7YAT6 A0A182IZ62 R4WND8 A0A0A0R243
Pubmed
26354079
22651552
14668217
19121390
17603110
22118469
+ More
26227816 24845553 29403074 26823975 25401762 20566863 21282665 20798317 21347285 24508170 18362917 19820115 28004739 30249741 21719571 20075255 23537049 22516182 26369729 25348373 25830018 27129103 18207082 17994087 15632085 25315136 17760985 23691247
26227816 24845553 29403074 26823975 25401762 20566863 21282665 20798317 21347285 24508170 18362917 19820115 28004739 30249741 21719571 20075255 23537049 22516182 26369729 25348373 25830018 27129103 18207082 17994087 15632085 25315136 17760985 23691247
EMBL
KQ460211
KPJ16499.1
NWSH01000372
PCG76868.1
AK402598
BAM19220.1
+ More
RSAL01000033 RVE51505.1 GDQN01004222 JAT86832.1 AK401165 BAM17787.1 AY769333 AAV34875.1 GU084287 ACY95320.1 AF400214 BABH01005868 KQ459585 KPI98359.1 EF207952 ABS57432.1 JF265048 AEL28879.1 ODYU01008712 SOQ52534.1 AGBW02010111 OWR49149.1 HQ424754 ADT80698.1 JTDY01004240 KOB68404.1 MG992447 AXY94885.1 AB180423 BAD26667.1 KK852463 KDR23421.1 UFQS01000164 UFQT01000164 UFQT01000683 SSX00665.1 SSX21045.1 SSX26537.1 PYGN01000102 PSN54288.1 AM048971 CAJ17206.1 NEVH01020350 PNF21374.1 GDHC01005447 JAQ13182.1 EF541358 ABQ18249.1 GBHO01004181 JAG39423.1 GU120454 ACY71304.1 DS234995 EEB10115.1 GDJX01023461 JAT44475.1 AJ783887 CAH04335.1 GL762454 EFZ21455.1 GL449036 EFN83368.1 KQ976401 KYM92411.1 GL444002 EFN61397.1 KQ977871 KYM99203.1 GU471222 ADC34228.1 ADTU01013450 GDHC01014916 JAQ03713.1 AJ783885 CAH04333.1 AJ783886 CAH04334.1 KQ981490 KYN41175.1 KK107489 EZA49877.1 KQ971352 EFA07317.1 GEZM01031312 JAV84940.1 LBMM01000198 KMR04507.1 QOIP01000010 RLU17698.1 GL888292 EGI63135.1 GEBQ01002931 JAT37046.1 QOIP01000008 RLU19644.1 AAZX01017041 GALX01000539 JAB67927.1 APGK01059439 KB741293 KB632046 ENN70273.1 ERL88282.1 DQ673392 ABG81965.1 GECZ01028231 GECZ01020002 JAS41538.1 JAS49767.1 GECU01029365 GECU01008645 JAS78341.1 JAS99061.1 GEDC01023252 JAS14046.1 KK107064 EZA60885.1 GFDL01010529 JAV24516.1 GANO01001048 JAB58823.1 DS231885 EDS43214.1 BT128367 AEE63325.1 GDAI01001204 JAI16399.1 GANO01001062 JAB58809.1 GBYB01002746 JAG72513.1 GAKP01005839 JAC53113.1 GBXI01010518 JAD03774.1 GDHF01018909 GDHF01017370 GDHF01016297 GDHF01006577 JAI33405.1 JAI34944.1 JAI36017.1 JAI45737.1 GDIQ01040739 GDIQ01004345 JAN53998.1 GFDF01004923 JAV09161.1 GDKW01000898 JAI55697.1 GECL01003698 JAP02426.1 ACPB03005019 GAHY01000891 JAA76619.1 EF638968 GEMB01000102 ABR27853.1 JAS03009.1 AJVK01023857 KM267674 AIU94800.1 CH963847 EDW73476.2 OUUW01000002 SPP77326.1 CH379067 EAL31355.2 CH479202 EDW30193.1 KQ434775 KZC04044.1 KQ435735 KOX77134.1 KZ288254 PBC30822.1 JR048093 AEY60575.1 EU032340 ABV44707.1 GFTR01002206 JAW14220.1 GFDG01001427 JAV17372.1 GBYB01003705 JAG73472.1 KQ435737 KOX76900.1 GFDG01001425 JAV17374.1 KQ414613 KOC69101.1 SPP77325.1 KC507314 AHB12437.1 AK417136 BAN20351.1 KM267684 AIU94810.1
RSAL01000033 RVE51505.1 GDQN01004222 JAT86832.1 AK401165 BAM17787.1 AY769333 AAV34875.1 GU084287 ACY95320.1 AF400214 BABH01005868 KQ459585 KPI98359.1 EF207952 ABS57432.1 JF265048 AEL28879.1 ODYU01008712 SOQ52534.1 AGBW02010111 OWR49149.1 HQ424754 ADT80698.1 JTDY01004240 KOB68404.1 MG992447 AXY94885.1 AB180423 BAD26667.1 KK852463 KDR23421.1 UFQS01000164 UFQT01000164 UFQT01000683 SSX00665.1 SSX21045.1 SSX26537.1 PYGN01000102 PSN54288.1 AM048971 CAJ17206.1 NEVH01020350 PNF21374.1 GDHC01005447 JAQ13182.1 EF541358 ABQ18249.1 GBHO01004181 JAG39423.1 GU120454 ACY71304.1 DS234995 EEB10115.1 GDJX01023461 JAT44475.1 AJ783887 CAH04335.1 GL762454 EFZ21455.1 GL449036 EFN83368.1 KQ976401 KYM92411.1 GL444002 EFN61397.1 KQ977871 KYM99203.1 GU471222 ADC34228.1 ADTU01013450 GDHC01014916 JAQ03713.1 AJ783885 CAH04333.1 AJ783886 CAH04334.1 KQ981490 KYN41175.1 KK107489 EZA49877.1 KQ971352 EFA07317.1 GEZM01031312 JAV84940.1 LBMM01000198 KMR04507.1 QOIP01000010 RLU17698.1 GL888292 EGI63135.1 GEBQ01002931 JAT37046.1 QOIP01000008 RLU19644.1 AAZX01017041 GALX01000539 JAB67927.1 APGK01059439 KB741293 KB632046 ENN70273.1 ERL88282.1 DQ673392 ABG81965.1 GECZ01028231 GECZ01020002 JAS41538.1 JAS49767.1 GECU01029365 GECU01008645 JAS78341.1 JAS99061.1 GEDC01023252 JAS14046.1 KK107064 EZA60885.1 GFDL01010529 JAV24516.1 GANO01001048 JAB58823.1 DS231885 EDS43214.1 BT128367 AEE63325.1 GDAI01001204 JAI16399.1 GANO01001062 JAB58809.1 GBYB01002746 JAG72513.1 GAKP01005839 JAC53113.1 GBXI01010518 JAD03774.1 GDHF01018909 GDHF01017370 GDHF01016297 GDHF01006577 JAI33405.1 JAI34944.1 JAI36017.1 JAI45737.1 GDIQ01040739 GDIQ01004345 JAN53998.1 GFDF01004923 JAV09161.1 GDKW01000898 JAI55697.1 GECL01003698 JAP02426.1 ACPB03005019 GAHY01000891 JAA76619.1 EF638968 GEMB01000102 ABR27853.1 JAS03009.1 AJVK01023857 KM267674 AIU94800.1 CH963847 EDW73476.2 OUUW01000002 SPP77326.1 CH379067 EAL31355.2 CH479202 EDW30193.1 KQ434775 KZC04044.1 KQ435735 KOX77134.1 KZ288254 PBC30822.1 JR048093 AEY60575.1 EU032340 ABV44707.1 GFTR01002206 JAW14220.1 GFDG01001427 JAV17372.1 GBYB01003705 JAG73472.1 KQ435737 KOX76900.1 GFDG01001425 JAV17374.1 KQ414613 KOC69101.1 SPP77325.1 KC507314 AHB12437.1 AK417136 BAN20351.1 KM267684 AIU94810.1
Proteomes
UP000053240
UP000218220
UP000283053
UP000005204
UP000053268
UP000007151
+ More
UP000037510 UP000027135 UP000245037 UP000235965 UP000009046 UP000192223 UP000008237 UP000078540 UP000000311 UP000078542 UP000005205 UP000078541 UP000053097 UP000007266 UP000036403 UP000279307 UP000007755 UP000002358 UP000019118 UP000030742 UP000002320 UP000095300 UP000015103 UP000092462 UP000007798 UP000268350 UP000001819 UP000008744 UP000076502 UP000053105 UP000242457 UP000005203 UP000095301 UP000053825 UP000075880
UP000037510 UP000027135 UP000245037 UP000235965 UP000009046 UP000192223 UP000008237 UP000078540 UP000000311 UP000078542 UP000005205 UP000078541 UP000053097 UP000007266 UP000036403 UP000279307 UP000007755 UP000002358 UP000019118 UP000030742 UP000002320 UP000095300 UP000015103 UP000092462 UP000007798 UP000268350 UP000001819 UP000008744 UP000076502 UP000053105 UP000242457 UP000005203 UP000095301 UP000053825 UP000075880
Pfam
PF00833 Ribosomal_S17e
SUPFAM
SSF116820
SSF116820
Gene 3D
ProteinModelPortal
A0A194RGC2
A0A2A4JYV9
I4DMT0
A0A437BMI5
A0A1E1WIM7
I4DIP7
+ More
Q5UAM5 D1LYL3 Q962R2 H9IW52 A0A194Q038 A7KCW4 G0ZEH0 A0A2H1WJA7 A0A212F614 E7DZ51 A0A0L7KZF2 A0A3G1T1N0 Q6F467 A0A067RHS7 A0A336LT32 A0A2P8ZCR8 Q4GXQ2 A0A2J7PYH7 A0A146M2H0 A5HMP2 A0A0A9Z7U0 D1M830 E0V9P6 A0A1D1XQ01 A0A1W4WSI4 Q6EUY4 E9ID71 E2BLN4 A0A195BWS5 E2AYV2 A0A151IEF9 D3KD00 A0A158NEI8 A0A146L5M2 Q6EUY6 Q6EUY5 A0A195FKI1 A0A026W1D6 D6WTW8 A0A1Y1MJZ0 A0A0J7L9E1 A0A3L8DCQ1 F4WRX8 A0A1B6MM82 A0A3L8DGT6 K7JAI6 V5H141 N6SYI9 Q0PXZ4 A0A1B6EUI2 A0A1B6HUL3 A0A1B6CKU2 A0A026WXT7 A0A1Q3FAB7 U5EXA4 B0WC62 J3JYN8 A0A0K8TPU7 U5EZC1 A0A1I8QF05 A0A0C9R7K5 A0A034WH81 A0A0A1WXJ6 A0A0K8V497 A0A0P6GVU4 A0A1L8DRY8 A0A0P4VME1 A0A0V0G4X0 R4G4C3 A6YPF7 A0A1B0D409 A0A0A0R236 B4MMS0 A0A3B0JVC5 Q29FA0 B4H1L6 A0A154NY80 A0A0N0BI35 A0A2A3EHS2 V9IHR8 A0A088ANM8 A0A1I8MSV1 A8C9W2 A0A224XZZ9 A0A1L8EF83 A0A0C9R6X9 A0A0M9A475 A0A1L8EFD0 A0A0L7REE3 A0A3B0J6C8 A0A0X7YAT6 A0A182IZ62 R4WND8 A0A0A0R243
Q5UAM5 D1LYL3 Q962R2 H9IW52 A0A194Q038 A7KCW4 G0ZEH0 A0A2H1WJA7 A0A212F614 E7DZ51 A0A0L7KZF2 A0A3G1T1N0 Q6F467 A0A067RHS7 A0A336LT32 A0A2P8ZCR8 Q4GXQ2 A0A2J7PYH7 A0A146M2H0 A5HMP2 A0A0A9Z7U0 D1M830 E0V9P6 A0A1D1XQ01 A0A1W4WSI4 Q6EUY4 E9ID71 E2BLN4 A0A195BWS5 E2AYV2 A0A151IEF9 D3KD00 A0A158NEI8 A0A146L5M2 Q6EUY6 Q6EUY5 A0A195FKI1 A0A026W1D6 D6WTW8 A0A1Y1MJZ0 A0A0J7L9E1 A0A3L8DCQ1 F4WRX8 A0A1B6MM82 A0A3L8DGT6 K7JAI6 V5H141 N6SYI9 Q0PXZ4 A0A1B6EUI2 A0A1B6HUL3 A0A1B6CKU2 A0A026WXT7 A0A1Q3FAB7 U5EXA4 B0WC62 J3JYN8 A0A0K8TPU7 U5EZC1 A0A1I8QF05 A0A0C9R7K5 A0A034WH81 A0A0A1WXJ6 A0A0K8V497 A0A0P6GVU4 A0A1L8DRY8 A0A0P4VME1 A0A0V0G4X0 R4G4C3 A6YPF7 A0A1B0D409 A0A0A0R236 B4MMS0 A0A3B0JVC5 Q29FA0 B4H1L6 A0A154NY80 A0A0N0BI35 A0A2A3EHS2 V9IHR8 A0A088ANM8 A0A1I8MSV1 A8C9W2 A0A224XZZ9 A0A1L8EF83 A0A0C9R6X9 A0A0M9A475 A0A1L8EFD0 A0A0L7REE3 A0A3B0J6C8 A0A0X7YAT6 A0A182IZ62 R4WND8 A0A0A0R243
PDB
4V6W
E-value=3.44444e-27,
Score=295
Ontologies
GO
PANTHER
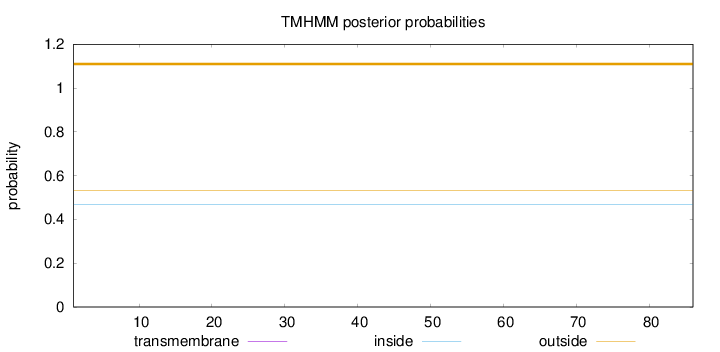
Topology
Length:
86
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00114
Exp number, first 60 AAs:
0
Total prob of N-in:
0.46799
outside
1 - 86
Population Genetic Test Statistics
Pi
192.342881
Theta
148.144169
Tajima's D
0.937812
CLR
0
CSRT
0.638918054097295
Interpretation
Uncertain
