Gene
KWMTBOMO12452 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002362
Annotation
cuticular_protein_RR-2_motif_75_precursor_[Bombyx_mori]
Full name
Cuticle protein
Location in the cell
Extracellular Reliability : 1.444 PlasmaMembrane Reliability : 1.067
Sequence
CDS
ATGGCAGCTAAGTTTTTCGTCGTCCTCTCCCTGGCGGTGGCCGCTTCAGCTCTTCCAGTTCTGGAATATGCTGAGCACGAAGCCCCCGCACACTACGACTTCGAGTACTCCGTTCACGACCAACAAAGCGGAGACATCAAGCAACAGAAGGAGTCCCGCGCCGGAGACGCCGTCCAGGGCTTCTACTCGCTGGTGCAACCCGACGGTGTCCACCGCATCGTCGAGTACACCTCCGACAAAGTAAACGGATTTAACGCTAACGTCCGCTACGAGGGACACCCCGTCGCTCAGCCCGCCAAGATCGCGTACGCCGCTCCCGTCGCCAAGCTCGCCTACTCCGCCCCCGTGGCCTATGCCGCACCCGTAGCCAAGATCGCCTACAGTGCTCCTGTTGCCAAGATCGCCTACAGCGCACCCATCGCCAAGGTCGCATACGCCGCCCCAGTTGCCAAGATCGCCTACAACAGTGCTCCCTTAACCCAAGTTACCTTCTCTTCCCCTGCCATTTCCTACCACCACTAG
Protein
MAAKFFVVLSLAVAASALPVLEYAEHEAPAHYDFEYSVHDQQSGDIKQQKESRAGDAVQGFYSLVQPDGVHRIVEYTSDKVNGFNANVRYEGHPVAQPAKIAYAAPVAKLAYSAPVAYAAPVAKIAYSAPVAKIAYSAPIAKVAYAAPVAKIAYNSAPLTQVTFSSPAISYHH
Summary
Keywords
Complete proteome
Cuticle
Reference proteome
Repeat
Feature
chain Cuticle protein
Uniprot
C0H6R7
C0H6Y0
C0H6R8
H9IW55
A0A437BLZ0
A0A3S2M4X4
+ More
C0H6R9 A0A1W5XG69 A0A194PYE1 B2DBI6 A0A2A4JZN9 A0A2A4JXY1 A0A212EX99 A0A2A4JZM8 A0A2A4JZQ5 A0A2A4JYI4 A0A212F3M1 A0A212FK22 I4DJ63 A0A194PZ85 A0A2A4JYJ5 I4DMC7 A0A212F1R9 A0A194Q033 B2DBI2 A0A194RKK4 A0A2H1WHI9 A0A194RGQ3 A0A194RGC7 A0A3S2NXG8 A0A2P8Y131 W5JUH9 A0A182VZH1 A0A1B0DNE8 A0A182VZG9 A0A2M4DMG7 A0A182ITA5 T1E9S8 Q17C02 A0A182H1C9 A0A084WBR6 A0A182W8E6 A0A182FQD0 A0A182LXP9 A0A3F2YV03 T1E733 A0A182FQD1 Q17015 A0A084VWT5 A0A1I8JTK7 A0A182XNI9 Q7Q0W8 A0A182WSM8 A0A336LLM9 A0A1W4WTD6 T1DES5 A0A182WSM6 W5JW36 A0A0L7QWA1 A0A182M9Z0 A0A1S4F8I9 A0A182H1C8 A0A084WBR8 B0X2J1 A0A182HLH8 A0A182XNI8 Q7PM04 A0A3F2YUT9 Q7PXZ2 A0A182HLH9 A0A1I8JTL5 A0A182L9W8
C0H6R9 A0A1W5XG69 A0A194PYE1 B2DBI6 A0A2A4JZN9 A0A2A4JXY1 A0A212EX99 A0A2A4JZM8 A0A2A4JZQ5 A0A2A4JYI4 A0A212F3M1 A0A212FK22 I4DJ63 A0A194PZ85 A0A2A4JYJ5 I4DMC7 A0A212F1R9 A0A194Q033 B2DBI2 A0A194RKK4 A0A2H1WHI9 A0A194RGQ3 A0A194RGC7 A0A3S2NXG8 A0A2P8Y131 W5JUH9 A0A182VZH1 A0A1B0DNE8 A0A182VZG9 A0A2M4DMG7 A0A182ITA5 T1E9S8 Q17C02 A0A182H1C9 A0A084WBR6 A0A182W8E6 A0A182FQD0 A0A182LXP9 A0A3F2YV03 T1E733 A0A182FQD1 Q17015 A0A084VWT5 A0A1I8JTK7 A0A182XNI9 Q7Q0W8 A0A182WSM8 A0A336LLM9 A0A1W4WTD6 T1DES5 A0A182WSM6 W5JW36 A0A0L7QWA1 A0A182M9Z0 A0A1S4F8I9 A0A182H1C8 A0A084WBR8 B0X2J1 A0A182HLH8 A0A182XNI8 Q7PM04 A0A3F2YUT9 Q7PXZ2 A0A182HLH9 A0A1I8JTL5 A0A182L9W8
Pubmed
EMBL
BABH01037875
BR000576
FAA00578.1
BR000639
FAA00641.1
BR000577
+ More
FAA00579.1 BABH01005857 RSAL01000033 RVE51500.1 RVE51499.1 BABH01005858 BR000578 FAA00580.1 KY554477 ARH56586.1 KQ459585 KPI98352.1 AB264666 BAG30745.1 NWSH01000372 PCG76880.1 PCG76881.1 AGBW02011788 OWR46128.1 PCG76870.1 PCG76882.1 PCG76869.1 AGBW02010555 OWR48324.1 AGBW02008159 OWR54088.1 AK401331 BAM17953.1 KPI98353.1 PCG76879.1 AK402445 BAM19067.1 AGBW02010835 OWR47664.1 KPI98354.1 AB264662 BAG30741.1 KQ460211 KPJ16506.1 ODYU01008712 SOQ52540.1 KPJ16505.1 KPJ16504.1 RVE51501.1 PYGN01001062 PSN37972.1 ADMH02000334 ETN66923.1 AJVK01017484 GGFL01014582 MBW78760.1 GAMD01001618 JAA99972.1 CH477314 EAT43817.1 JXUM01103160 KQ564781 KXJ71758.1 ATLV01022426 KE525332 KFB47660.1 AXCM01013007 GAMD01003387 JAA98203.1 AAAB01008987 U50469 ATLV01017753 KE525185 KFB42429.1 APCN01001621 AAAB01008980 EAA14336.2 UFQT01000011 SSX17569.1 GAMD01003411 JAA98179.1 ADMH02000274 ETN67159.1 KQ414716 KOC62887.1 AXCM01017960 KXJ71757.1 ATLV01022427 KFB47662.1 DS232291 EDS39214.1 EDS39215.1 APCN01000904 EAA14555.3 EAU76101.1 EAA01340.3 EAL38557.2
FAA00579.1 BABH01005857 RSAL01000033 RVE51500.1 RVE51499.1 BABH01005858 BR000578 FAA00580.1 KY554477 ARH56586.1 KQ459585 KPI98352.1 AB264666 BAG30745.1 NWSH01000372 PCG76880.1 PCG76881.1 AGBW02011788 OWR46128.1 PCG76870.1 PCG76882.1 PCG76869.1 AGBW02010555 OWR48324.1 AGBW02008159 OWR54088.1 AK401331 BAM17953.1 KPI98353.1 PCG76879.1 AK402445 BAM19067.1 AGBW02010835 OWR47664.1 KPI98354.1 AB264662 BAG30741.1 KQ460211 KPJ16506.1 ODYU01008712 SOQ52540.1 KPJ16505.1 KPJ16504.1 RVE51501.1 PYGN01001062 PSN37972.1 ADMH02000334 ETN66923.1 AJVK01017484 GGFL01014582 MBW78760.1 GAMD01001618 JAA99972.1 CH477314 EAT43817.1 JXUM01103160 KQ564781 KXJ71758.1 ATLV01022426 KE525332 KFB47660.1 AXCM01013007 GAMD01003387 JAA98203.1 AAAB01008987 U50469 ATLV01017753 KE525185 KFB42429.1 APCN01001621 AAAB01008980 EAA14336.2 UFQT01000011 SSX17569.1 GAMD01003411 JAA98179.1 ADMH02000274 ETN67159.1 KQ414716 KOC62887.1 AXCM01017960 KXJ71757.1 ATLV01022427 KFB47662.1 DS232291 EDS39214.1 EDS39215.1 APCN01000904 EAA14555.3 EAU76101.1 EAA01340.3 EAL38557.2
Proteomes
Interpro
IPR000618
Insect_cuticle
+ More
IPR031311 CHIT_BIND_RR_consensus
IPR002557 Chitin-bd_dom
IPR036508 Chitin-bd_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR011044 Quino_amine_DH_bsu
IPR011045 N2O_reductase_N
IPR031311 CHIT_BIND_RR_consensus
IPR002557 Chitin-bd_dom
IPR036508 Chitin-bd_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR011044 Quino_amine_DH_bsu
IPR011045 N2O_reductase_N
Gene 3D
ProteinModelPortal
C0H6R7
C0H6Y0
C0H6R8
H9IW55
A0A437BLZ0
A0A3S2M4X4
+ More
C0H6R9 A0A1W5XG69 A0A194PYE1 B2DBI6 A0A2A4JZN9 A0A2A4JXY1 A0A212EX99 A0A2A4JZM8 A0A2A4JZQ5 A0A2A4JYI4 A0A212F3M1 A0A212FK22 I4DJ63 A0A194PZ85 A0A2A4JYJ5 I4DMC7 A0A212F1R9 A0A194Q033 B2DBI2 A0A194RKK4 A0A2H1WHI9 A0A194RGQ3 A0A194RGC7 A0A3S2NXG8 A0A2P8Y131 W5JUH9 A0A182VZH1 A0A1B0DNE8 A0A182VZG9 A0A2M4DMG7 A0A182ITA5 T1E9S8 Q17C02 A0A182H1C9 A0A084WBR6 A0A182W8E6 A0A182FQD0 A0A182LXP9 A0A3F2YV03 T1E733 A0A182FQD1 Q17015 A0A084VWT5 A0A1I8JTK7 A0A182XNI9 Q7Q0W8 A0A182WSM8 A0A336LLM9 A0A1W4WTD6 T1DES5 A0A182WSM6 W5JW36 A0A0L7QWA1 A0A182M9Z0 A0A1S4F8I9 A0A182H1C8 A0A084WBR8 B0X2J1 A0A182HLH8 A0A182XNI8 Q7PM04 A0A3F2YUT9 Q7PXZ2 A0A182HLH9 A0A1I8JTL5 A0A182L9W8
C0H6R9 A0A1W5XG69 A0A194PYE1 B2DBI6 A0A2A4JZN9 A0A2A4JXY1 A0A212EX99 A0A2A4JZM8 A0A2A4JZQ5 A0A2A4JYI4 A0A212F3M1 A0A212FK22 I4DJ63 A0A194PZ85 A0A2A4JYJ5 I4DMC7 A0A212F1R9 A0A194Q033 B2DBI2 A0A194RKK4 A0A2H1WHI9 A0A194RGQ3 A0A194RGC7 A0A3S2NXG8 A0A2P8Y131 W5JUH9 A0A182VZH1 A0A1B0DNE8 A0A182VZG9 A0A2M4DMG7 A0A182ITA5 T1E9S8 Q17C02 A0A182H1C9 A0A084WBR6 A0A182W8E6 A0A182FQD0 A0A182LXP9 A0A3F2YV03 T1E733 A0A182FQD1 Q17015 A0A084VWT5 A0A1I8JTK7 A0A182XNI9 Q7Q0W8 A0A182WSM8 A0A336LLM9 A0A1W4WTD6 T1DES5 A0A182WSM6 W5JW36 A0A0L7QWA1 A0A182M9Z0 A0A1S4F8I9 A0A182H1C8 A0A084WBR8 B0X2J1 A0A182HLH8 A0A182XNI8 Q7PM04 A0A3F2YUT9 Q7PXZ2 A0A182HLH9 A0A1I8JTL5 A0A182L9W8
Ontologies
GO
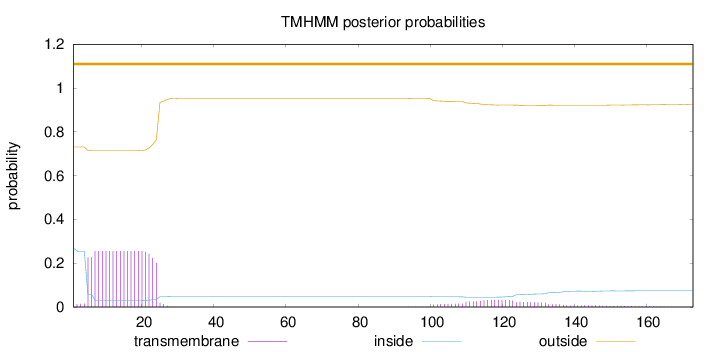
Topology
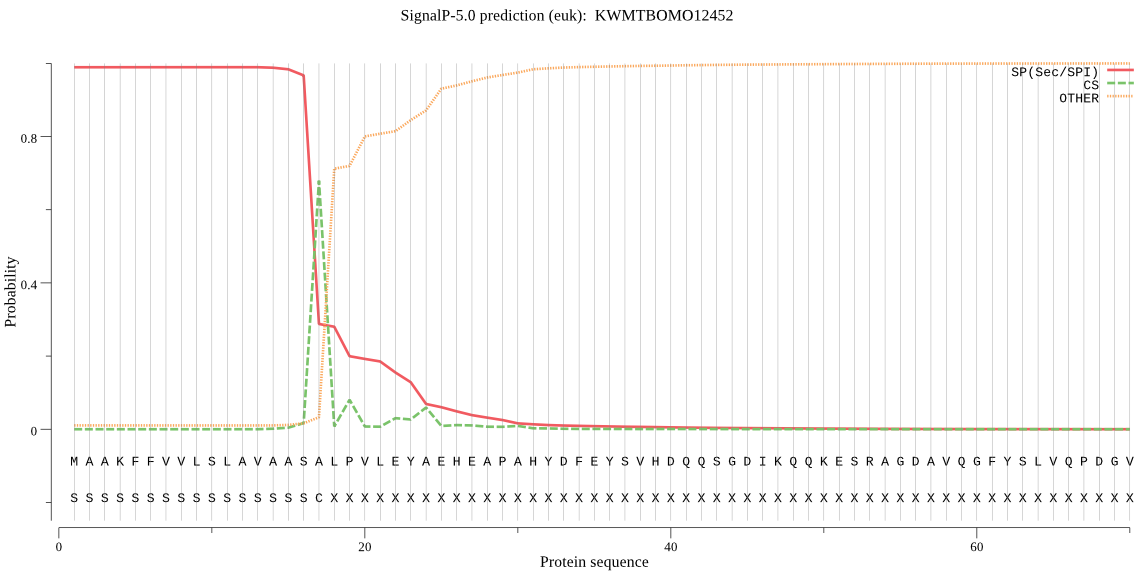
SignalP
Position: 1 - 17,
Likelihood: 0.989120
Length:
173
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.95809999999999
Exp number, first 60 AAs:
5.03396
Total prob of N-in:
0.26883
outside
1 - 173
Population Genetic Test Statistics
Pi
132.700837
Theta
153.382076
Tajima's D
-0.400562
CLR
42.475913
CSRT
0.265586720663967
Interpretation
Uncertain
