Pre Gene Modal
BGIBMGA002363
Annotation
PREDICTED:_pre-mRNA-splicing_factor_RBM22_isoform_X1_[Amyelois_transitella]
Full name
Pre-mRNA-splicing factor RBM22
Alternative Name
RNA-binding motif protein 22
Location in the cell
Nuclear Reliability : 3.319
Sequence
CDS
ATGGCTACATCGAAGTCAACAAACACATACAATCGTCAGAATTGGGAAGACGCCGATTTTCCAATATTGTGCCAAACATGTCTTGGAGATAACCCTTACATTAGAATGACTAAAGAAAAATACGGTAAAGAATGTAAAATATGCTCAAGACCTTTCACAGTGTTTCGATGGTGTCCCGGAGCAAGAATGCGTTTCAAGAAAACAGAAATATGTCAGACCTGTTCGAAACTGAAAAATGTTTGTCAAACATGTTTATTGGATTTGGAATATGGTCTGCCAATCCAAGTGAGAGATGCTGCATTAAAAGTTCAGGATGACCTACCTCGAAACGAAGTAAACAAGGAATATTACATACAAAACTTGGAGAGTCAGCTGTCTAACTCTGATCCAACACAGCCAACCAATTCTCTAAAATCTAAAGGATCGTCCGATTTGTTGGTGCGGTTAGCTAGAACAGCTCCATATTATAAGAGGAATAGACCGCACGTGTGCTCGTTTTGGGTCAAAGGAGAATGTAGACGAGGGGAAGAATGTCCGTATCGTCACGAGAAGCCAACAGATCCTGATGATCCACTAGCTGATCAGAACATTAAAGACAGGTATTATGGTGTAAATGATCCAGTAGCTGAGAAGCTTATGCGTCGTGCTGCTGCAATGCCTGCTCTACCTCCACCTGAAGACAAAACTGTTACTACCTTGTATGTTGGTAATCTTCCTGATAATGTCACTGAGGACGAGCTTAGAGGCCACTTTTACCAATATGGTGAAATTAGATGTTTGACTCTTGTACCTCGAGCCCAGTGTGCCTTTGTGCAATATACTTCAAGAAATGCTGCAGAACATGCTGCAGAAAAGACTTTCAATCGTCTTGTTATTTTTGGCAAGAGACTGACAATCAAATGGGGTAAATCGCAAGGACGACAAGGACTTACTGAGAAGAATGATGGTGTACCCATGTTGGAACCAGTTCCTGGATTGCCTGGGACGTTGCCACCACCCCCTGCATATTTACATCCATTCCCGCCACAAATGCCACCACCCCCGAACCGTCCGAACGATTTCTTCAACTTGCACCACTATGGTGGTCCGGGCTGGGGACCGTGGGGTCCGCCTGGCATGGCGCCGCCCCCGCCTCCGGCTCCGCTCCCGGCCCCGCTCCGCCTGCACTACCCTAGCCAGGACCCCGCGCGTCTCGGGGCCCACGCACACACCAACGCGCCGCAAATTTAG
Protein
MATSKSTNTYNRQNWEDADFPILCQTCLGDNPYIRMTKEKYGKECKICSRPFTVFRWCPGARMRFKKTEICQTCSKLKNVCQTCLLDLEYGLPIQVRDAALKVQDDLPRNEVNKEYYIQNLESQLSNSDPTQPTNSLKSKGSSDLLVRLARTAPYYKRNRPHVCSFWVKGECRRGEECPYRHEKPTDPDDPLADQNIKDRYYGVNDPVAEKLMRRAAAMPALPPPEDKTVTTLYVGNLPDNVTEDELRGHFYQYGEIRCLTLVPRAQCAFVQYTSRNAAEHAAEKTFNRLVIFGKRLTIKWGKSQGRQGLTEKNDGVPMLEPVPGLPGTLPPPPAYLHPFPPQMPPPPNRPNDFFNLHHYGGPGWGPWGPPGMAPPPPPAPLPAPLRLHYPSQDPARLGAHAHTNAPQI
Summary
Description
Required for pre-mRNA splicing as component of the activated spliceosome. Involved in the first step of pre-mRNA splicing. Binds directly to the internal stem-loop (ISL) domain of the U6 snRNA and to the pre-mRNA intron near the 5' splice site during the activation and catalytic phases of the spliceosome cycle.
Subunit
Component of the pre-catalytic and catalytic spliceosome complexes. Component of the postcatalytic spliceosome P complex.
Similarity
Belongs to the SLT11 family.
Keywords
Complete proteome
Cytoplasm
Developmental protein
Metal-binding
mRNA processing
mRNA splicing
Nucleus
Reference proteome
RNA-binding
Spliceosome
Zinc
Zinc-finger
Feature
chain Pre-mRNA-splicing factor RBM22
Uniprot
H9IYM9
A0A2A4JZD6
A0A437BLZ6
A0A2H1W9Y3
A0A194RFX9
A0A194Q028
+ More
S4NSX0 A0A212FK43 A0A0L7LIH4 A0A2P8Z4J5 A0A2J7Q028 A0A0L7QQG2 A0A1B6ME92 A0A1B6I9I3 E2ABI2 A0A1B6F802 A0A310SFR3 A0A026WDM4 A0A195DV40 A0A2A3E1N9 A0A088AA93 A0A154PFY2 T1HSL9 A0A151I648 A0A158NSN0 A0A195EWS4 U4UP50 A0A1Y1N1G3 A0A0A9WE08 A0A023F4S6 A0A224XBU7 A0A0P4VVH9 A0A0V0G8K8 A0A069DUC6 K7IV25 E9ISD3 A0A0C9RMA2 A0A3P8VR60 A0A1A7WFT3 E2C802 A0A3B4DXG8 G9KKG3 A0A3Q2ZQ23 A0A0D9SCD2 A0A1A8S5V8 A0A1A8QKL7 A0A1A8DU49 A0A1A8UF43 Q6P616 A0A2K6V3K5 A0A1A8I096 A0A2I4B7K1 A0A2K5EFV4 A0A2K5RS79 U3DF69 H2LJT2 A0A3P9JU52 A0A0S7LE07 A0A3B3BEC6 A0A341ADM5 A0A3Q7Q430 A0A2D0SY08 A0A1S3CVF3 A0A0P6JBH5 A0A1U7QIN5 A0A250Y0K3 H0UXE0 A0A2K6CJJ1 A0A2K6MK02 G1T1G7 G1PCS9 G3RAZ8 G1LD30 G1RHG3 A0A2K5Z0B5 A0A452ESB3 G3SYJ2 A0A2U3V6I4 A0A3Q7NMY5 A0A340WLW8 A0A2R9B305 A0A096N735 A0A2Y9FAQ6 A0A2K6R5N5 A0A2Y9HM80 M3WPJ5 A0A2K5NNP0 A0A2Y9ISK1 A0A3Q7TSP1 A0A3P4LT42 A0A2Y9PSH0 A0A1U7TVT4 F2Z4G3 M3YQP8 A0A2K5HTL7 A0A384AMW6 A0A452SKA6 A0A3Q7W055 H2PH39 A0A384C7D3
S4NSX0 A0A212FK43 A0A0L7LIH4 A0A2P8Z4J5 A0A2J7Q028 A0A0L7QQG2 A0A1B6ME92 A0A1B6I9I3 E2ABI2 A0A1B6F802 A0A310SFR3 A0A026WDM4 A0A195DV40 A0A2A3E1N9 A0A088AA93 A0A154PFY2 T1HSL9 A0A151I648 A0A158NSN0 A0A195EWS4 U4UP50 A0A1Y1N1G3 A0A0A9WE08 A0A023F4S6 A0A224XBU7 A0A0P4VVH9 A0A0V0G8K8 A0A069DUC6 K7IV25 E9ISD3 A0A0C9RMA2 A0A3P8VR60 A0A1A7WFT3 E2C802 A0A3B4DXG8 G9KKG3 A0A3Q2ZQ23 A0A0D9SCD2 A0A1A8S5V8 A0A1A8QKL7 A0A1A8DU49 A0A1A8UF43 Q6P616 A0A2K6V3K5 A0A1A8I096 A0A2I4B7K1 A0A2K5EFV4 A0A2K5RS79 U3DF69 H2LJT2 A0A3P9JU52 A0A0S7LE07 A0A3B3BEC6 A0A341ADM5 A0A3Q7Q430 A0A2D0SY08 A0A1S3CVF3 A0A0P6JBH5 A0A1U7QIN5 A0A250Y0K3 H0UXE0 A0A2K6CJJ1 A0A2K6MK02 G1T1G7 G1PCS9 G3RAZ8 G1LD30 G1RHG3 A0A2K5Z0B5 A0A452ESB3 G3SYJ2 A0A2U3V6I4 A0A3Q7NMY5 A0A340WLW8 A0A2R9B305 A0A096N735 A0A2Y9FAQ6 A0A2K6R5N5 A0A2Y9HM80 M3WPJ5 A0A2K5NNP0 A0A2Y9ISK1 A0A3Q7TSP1 A0A3P4LT42 A0A2Y9PSH0 A0A1U7TVT4 F2Z4G3 M3YQP8 A0A2K5HTL7 A0A384AMW6 A0A452SKA6 A0A3Q7W055 H2PH39 A0A384C7D3
Pubmed
19121390
26354079
23622113
22118469
26227816
29403074
+ More
20798317 24508170 30249741 21347285 23537049 28004739 25401762 26823975 25474469 27129103 26334808 20075255 21282665 24487278 23236062 25243066 17554307 29451363 28087693 21993624 22398555 20010809 22722832 25362486 17975172 19393038 24813606
20798317 24508170 30249741 21347285 23537049 28004739 25401762 26823975 25474469 27129103 26334808 20075255 21282665 24487278 23236062 25243066 17554307 29451363 28087693 21993624 22398555 20010809 22722832 25362486 17975172 19393038 24813606
EMBL
BABH01037876
NWSH01000372
PCG76873.1
RSAL01000033
RVE51496.1
ODYU01007251
+ More
SOQ49863.1 KQ460211 KPJ16507.1 KQ459585 KPI98349.1 GAIX01013872 JAA78688.1 AGBW02008159 OWR54086.1 JTDY01001003 KOB75144.1 PYGN01000199 PSN51415.1 NEVH01020327 PNF21931.1 KQ414786 KOC60857.1 GEBQ01005752 JAT34225.1 GECU01024100 JAS83606.1 GL438255 EFN69209.1 GECZ01023434 JAS46335.1 KQ765605 OAD53866.1 KK107260 QOIP01000001 EZA54063.1 RLU26895.1 KQ980322 KYN16612.1 KZ288466 PBC25444.1 KQ434896 KZC10732.1 ACPB03008326 KQ976400 KYM92661.1 ADTU01025062 ADTU01025063 KQ981948 KYN32601.1 KB632308 ERL91926.1 GEZM01015396 JAV91689.1 GBHO01038841 GBHO01038839 GBRD01002990 GDHC01002015 JAG04763.1 JAG04765.1 JAG62831.1 JAQ16614.1 GBBI01002733 JAC15979.1 GFTR01006561 JAW09865.1 GDKW01000010 JAI56585.1 GECL01001766 JAP04358.1 GBGD01001334 JAC87555.1 AAZX01001182 GL765332 EFZ16520.1 GBYB01008106 JAG77873.1 HADW01003442 HADX01015570 SBP04842.1 GL453506 EFN75920.1 JP016794 AES05392.1 AQIB01086595 HAEH01018030 HAEI01011551 SBS13641.1 HAEF01020224 HAEG01013082 SBR94007.1 HADZ01012423 HAEA01007874 SBQ36354.1 HADY01016083 HAEJ01006263 SBS46720.1 CR761977 BC062518 HAED01005064 HAEE01011240 SBQ91094.1 GAMS01011110 GAMR01007493 GAMQ01005997 GAMP01009972 JAB12026.1 JAB26439.1 JAB35854.1 JAB42783.1 GBYX01174170 JAO87661.1 GEBF01001980 JAO01653.1 GFFV01002831 JAV37114.1 AAKN02010073 AAGW02027421 AAGW02027422 AAPE02018683 CABD030041695 ACTA01096661 ADFV01033842 LWLT01000008 AJFE02102361 AHZZ02030202 AANG04003171 CYRY02002257 VCW66914.1 AEYP01000949 ABGA01017759 ABGA01017760 NDHI03003371 PNJ77512.1
SOQ49863.1 KQ460211 KPJ16507.1 KQ459585 KPI98349.1 GAIX01013872 JAA78688.1 AGBW02008159 OWR54086.1 JTDY01001003 KOB75144.1 PYGN01000199 PSN51415.1 NEVH01020327 PNF21931.1 KQ414786 KOC60857.1 GEBQ01005752 JAT34225.1 GECU01024100 JAS83606.1 GL438255 EFN69209.1 GECZ01023434 JAS46335.1 KQ765605 OAD53866.1 KK107260 QOIP01000001 EZA54063.1 RLU26895.1 KQ980322 KYN16612.1 KZ288466 PBC25444.1 KQ434896 KZC10732.1 ACPB03008326 KQ976400 KYM92661.1 ADTU01025062 ADTU01025063 KQ981948 KYN32601.1 KB632308 ERL91926.1 GEZM01015396 JAV91689.1 GBHO01038841 GBHO01038839 GBRD01002990 GDHC01002015 JAG04763.1 JAG04765.1 JAG62831.1 JAQ16614.1 GBBI01002733 JAC15979.1 GFTR01006561 JAW09865.1 GDKW01000010 JAI56585.1 GECL01001766 JAP04358.1 GBGD01001334 JAC87555.1 AAZX01001182 GL765332 EFZ16520.1 GBYB01008106 JAG77873.1 HADW01003442 HADX01015570 SBP04842.1 GL453506 EFN75920.1 JP016794 AES05392.1 AQIB01086595 HAEH01018030 HAEI01011551 SBS13641.1 HAEF01020224 HAEG01013082 SBR94007.1 HADZ01012423 HAEA01007874 SBQ36354.1 HADY01016083 HAEJ01006263 SBS46720.1 CR761977 BC062518 HAED01005064 HAEE01011240 SBQ91094.1 GAMS01011110 GAMR01007493 GAMQ01005997 GAMP01009972 JAB12026.1 JAB26439.1 JAB35854.1 JAB42783.1 GBYX01174170 JAO87661.1 GEBF01001980 JAO01653.1 GFFV01002831 JAV37114.1 AAKN02010073 AAGW02027421 AAGW02027422 AAPE02018683 CABD030041695 ACTA01096661 ADFV01033842 LWLT01000008 AJFE02102361 AHZZ02030202 AANG04003171 CYRY02002257 VCW66914.1 AEYP01000949 ABGA01017759 ABGA01017760 NDHI03003371 PNJ77512.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000245037 UP000235965 UP000053825 UP000000311 UP000053097 UP000279307 UP000078492 UP000242457 UP000005203 UP000076502 UP000015103 UP000078540 UP000005205 UP000078541 UP000030742 UP000002358 UP000265120 UP000008237 UP000261440 UP000264800 UP000029965 UP000008143 UP000233220 UP000192220 UP000233020 UP000233040 UP000008225 UP000001038 UP000265180 UP000265200 UP000261560 UP000252040 UP000286641 UP000221080 UP000079169 UP000189706 UP000005447 UP000233120 UP000233180 UP000001811 UP000001074 UP000001519 UP000008912 UP000001073 UP000233140 UP000291000 UP000007646 UP000245320 UP000265300 UP000240080 UP000028761 UP000233200 UP000248481 UP000011712 UP000233060 UP000248482 UP000286640 UP000248483 UP000189704 UP000009136 UP000000715 UP000233080 UP000261681 UP000291022 UP000286642 UP000001595 UP000261680 UP000291021
UP000037510 UP000245037 UP000235965 UP000053825 UP000000311 UP000053097 UP000279307 UP000078492 UP000242457 UP000005203 UP000076502 UP000015103 UP000078540 UP000005205 UP000078541 UP000030742 UP000002358 UP000265120 UP000008237 UP000261440 UP000264800 UP000029965 UP000008143 UP000233220 UP000192220 UP000233020 UP000233040 UP000008225 UP000001038 UP000265180 UP000265200 UP000261560 UP000252040 UP000286641 UP000221080 UP000079169 UP000189706 UP000005447 UP000233120 UP000233180 UP000001811 UP000001074 UP000001519 UP000008912 UP000001073 UP000233140 UP000291000 UP000007646 UP000245320 UP000265300 UP000240080 UP000028761 UP000233200 UP000248481 UP000011712 UP000233060 UP000248482 UP000286640 UP000248483 UP000189704 UP000009136 UP000000715 UP000233080 UP000261681 UP000291022 UP000286642 UP000001595 UP000261680 UP000291021
Pfam
PF00076 RRM_1
Interpro
Gene 3D
ProteinModelPortal
H9IYM9
A0A2A4JZD6
A0A437BLZ6
A0A2H1W9Y3
A0A194RFX9
A0A194Q028
+ More
S4NSX0 A0A212FK43 A0A0L7LIH4 A0A2P8Z4J5 A0A2J7Q028 A0A0L7QQG2 A0A1B6ME92 A0A1B6I9I3 E2ABI2 A0A1B6F802 A0A310SFR3 A0A026WDM4 A0A195DV40 A0A2A3E1N9 A0A088AA93 A0A154PFY2 T1HSL9 A0A151I648 A0A158NSN0 A0A195EWS4 U4UP50 A0A1Y1N1G3 A0A0A9WE08 A0A023F4S6 A0A224XBU7 A0A0P4VVH9 A0A0V0G8K8 A0A069DUC6 K7IV25 E9ISD3 A0A0C9RMA2 A0A3P8VR60 A0A1A7WFT3 E2C802 A0A3B4DXG8 G9KKG3 A0A3Q2ZQ23 A0A0D9SCD2 A0A1A8S5V8 A0A1A8QKL7 A0A1A8DU49 A0A1A8UF43 Q6P616 A0A2K6V3K5 A0A1A8I096 A0A2I4B7K1 A0A2K5EFV4 A0A2K5RS79 U3DF69 H2LJT2 A0A3P9JU52 A0A0S7LE07 A0A3B3BEC6 A0A341ADM5 A0A3Q7Q430 A0A2D0SY08 A0A1S3CVF3 A0A0P6JBH5 A0A1U7QIN5 A0A250Y0K3 H0UXE0 A0A2K6CJJ1 A0A2K6MK02 G1T1G7 G1PCS9 G3RAZ8 G1LD30 G1RHG3 A0A2K5Z0B5 A0A452ESB3 G3SYJ2 A0A2U3V6I4 A0A3Q7NMY5 A0A340WLW8 A0A2R9B305 A0A096N735 A0A2Y9FAQ6 A0A2K6R5N5 A0A2Y9HM80 M3WPJ5 A0A2K5NNP0 A0A2Y9ISK1 A0A3Q7TSP1 A0A3P4LT42 A0A2Y9PSH0 A0A1U7TVT4 F2Z4G3 M3YQP8 A0A2K5HTL7 A0A384AMW6 A0A452SKA6 A0A3Q7W055 H2PH39 A0A384C7D3
S4NSX0 A0A212FK43 A0A0L7LIH4 A0A2P8Z4J5 A0A2J7Q028 A0A0L7QQG2 A0A1B6ME92 A0A1B6I9I3 E2ABI2 A0A1B6F802 A0A310SFR3 A0A026WDM4 A0A195DV40 A0A2A3E1N9 A0A088AA93 A0A154PFY2 T1HSL9 A0A151I648 A0A158NSN0 A0A195EWS4 U4UP50 A0A1Y1N1G3 A0A0A9WE08 A0A023F4S6 A0A224XBU7 A0A0P4VVH9 A0A0V0G8K8 A0A069DUC6 K7IV25 E9ISD3 A0A0C9RMA2 A0A3P8VR60 A0A1A7WFT3 E2C802 A0A3B4DXG8 G9KKG3 A0A3Q2ZQ23 A0A0D9SCD2 A0A1A8S5V8 A0A1A8QKL7 A0A1A8DU49 A0A1A8UF43 Q6P616 A0A2K6V3K5 A0A1A8I096 A0A2I4B7K1 A0A2K5EFV4 A0A2K5RS79 U3DF69 H2LJT2 A0A3P9JU52 A0A0S7LE07 A0A3B3BEC6 A0A341ADM5 A0A3Q7Q430 A0A2D0SY08 A0A1S3CVF3 A0A0P6JBH5 A0A1U7QIN5 A0A250Y0K3 H0UXE0 A0A2K6CJJ1 A0A2K6MK02 G1T1G7 G1PCS9 G3RAZ8 G1LD30 G1RHG3 A0A2K5Z0B5 A0A452ESB3 G3SYJ2 A0A2U3V6I4 A0A3Q7NMY5 A0A340WLW8 A0A2R9B305 A0A096N735 A0A2Y9FAQ6 A0A2K6R5N5 A0A2Y9HM80 M3WPJ5 A0A2K5NNP0 A0A2Y9ISK1 A0A3Q7TSP1 A0A3P4LT42 A0A2Y9PSH0 A0A1U7TVT4 F2Z4G3 M3YQP8 A0A2K5HTL7 A0A384AMW6 A0A452SKA6 A0A3Q7W055 H2PH39 A0A384C7D3
PDB
6ID1
E-value=7.39787e-140,
Score=1274
Ontologies
KEGG
GO
PANTHER
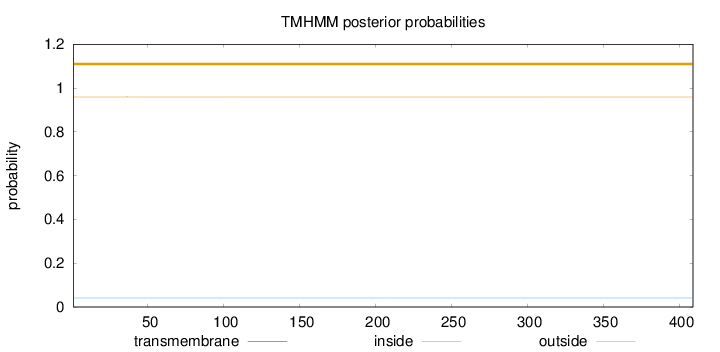
Topology
Subcellular location
Nucleus
Nearly exclusively nuclear. May be shuttling between the nucleus and the cytosol. With evidence from 2 publications.
Cytoplasm Nearly exclusively nuclear. May be shuttling between the nucleus and the cytosol. With evidence from 2 publications.
Cytoplasm Nearly exclusively nuclear. May be shuttling between the nucleus and the cytosol. With evidence from 2 publications.
Length:
409
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000660000000000001
Exp number, first 60 AAs:
0.00051
Total prob of N-in:
0.04058
outside
1 - 409
Population Genetic Test Statistics
Pi
225.147958
Theta
191.320647
Tajima's D
0.27331
CLR
0
CSRT
0.451127443627819
Interpretation
Uncertain
