Gene
KWMTBOMO12444
Annotation
mariner_transposase_[Bombyx_mori]
Full name
Innexin
Location in the cell
Cytoplasmic Reliability : 1.307 Nuclear Reliability : 1.659
Sequence
CDS
ATGGTGGCCACGTTTGTCTCCAAAACAGGCCATGTTACGACTATTCCTCTTGAGGGACAAGGAACGGTTAATGCAGAATGGTATGCTAGCATTTGTTTGCCACAGGTCGTTTCTGAACTCCGTAAGGAGAACTGCAACCGCCGCATCATCCTCCATCACGACAATGCGAGTTCTCACACCGCGCACAGAACAAAAGAGTTTTTAGAGCAAGAAAACATAGAATTATTAGACCATCCGCCGTACAGCCCCGACCTAAGCCCTAATGATTTCTATACTTTCCCTAAAATAAAGAATAAATTACGTGGACAGAGATTTTCATCACCTGAAGAAGCTGTGGACGTCTACAAAACGGCCATTTTGGAGACCCCAACTTCCGAATGGAATGGTTGCTTCAATGATTGGCTCCGTCGTATGGAAAAATGTGTCAAATTTCGCGGAGAATACTTCGAAAAGCAATAA
Protein
MVATFVSKTGHVTTIPLEGQGTVNAEWYASICLPQVVSELRKENCNRRIILHHDNASSHTAHRTKEFLEQENIELLDHPPYSPDLSPNDFYTFPKIKNKLRGQRFSSPEEAVDVYKTAILETPTSEWNGCFNDWLRRMEKCVKFRGEYFEKQ
Summary
Description
Structural component of the gap junctions.
Similarity
Belongs to the pannexin family.
Uniprot
Q1HPJ3
A0A0J7K8F0
A0A1B6G5C6
A0A034VN11
A0A1L5BY10
A0A069DT60
+ More
A0A023F6H6 A0A0V0G5J0 A0A0K8U1A4 A0A0K2VK65 A0A267ESW3 A0A1I8HNK7 F4W604 A0A0K2VDK7 A0A267EV83 A0A267G4K5 Q0QXC1 B3CK18 A6GV71 B7ZDI8 A6GV70 A6GV69 B7ZDI6 A6GV72 B7ZDI7 B3CK19 A0A1B0CCE3 B3CK24 B3CK26 B3CK17 A0A1W0X0B6 B3CK30 B3CK29 B3CK23 A0A1B6JDU2 A0A034V2R5 Q13539 A0A1W0WCL5 A0A2H9T3D9 A0A2S2QMC5 A0A2S2RAU4 A0A2J7R7K0 A0A1W0WPR3 A0A023F6W4 A0A1L5BY05 X1X2D9 A0A0J7K1H8 A0A023F6P2 A0A2J7Q9R7 A0A1B6MR58 A0A2S2Q0U2 A0A151JAL8 A0A2P6KLU2 A0A2J7R3C8 A0A224XTA3 A0A2J7QND6 A0A2J7RS37 E2ADI9 A0A1L5BY09 J9LNR4 A0A3Q2GQF1 A0A0J7KB73 A0A0K2UGY4 A0A0J7JXI1 A0A1L5BXZ8 A0A1B6HD45 A0A1I8HRU7 A0A2J7PE56 A0A1B6F2S5
A0A023F6H6 A0A0V0G5J0 A0A0K8U1A4 A0A0K2VK65 A0A267ESW3 A0A1I8HNK7 F4W604 A0A0K2VDK7 A0A267EV83 A0A267G4K5 Q0QXC1 B3CK18 A6GV71 B7ZDI8 A6GV70 A6GV69 B7ZDI6 A6GV72 B7ZDI7 B3CK19 A0A1B0CCE3 B3CK24 B3CK26 B3CK17 A0A1W0X0B6 B3CK30 B3CK29 B3CK23 A0A1B6JDU2 A0A034V2R5 Q13539 A0A1W0WCL5 A0A2H9T3D9 A0A2S2QMC5 A0A2S2RAU4 A0A2J7R7K0 A0A1W0WPR3 A0A023F6W4 A0A1L5BY05 X1X2D9 A0A0J7K1H8 A0A023F6P2 A0A2J7Q9R7 A0A1B6MR58 A0A2S2Q0U2 A0A151JAL8 A0A2P6KLU2 A0A2J7R3C8 A0A224XTA3 A0A2J7QND6 A0A2J7RS37 E2ADI9 A0A1L5BY09 J9LNR4 A0A3Q2GQF1 A0A0J7KB73 A0A0K2UGY4 A0A0J7JXI1 A0A1L5BXZ8 A0A1B6HD45 A0A1I8HRU7 A0A2J7PE56 A0A1B6F2S5
Pubmed
EMBL
DQ443409
ABF51498.1
LBMM01012006
KMQ86491.1
GECZ01012256
JAS57513.1
+ More
GAKP01015435 JAC43517.1 KX930998 APL98291.1 GBGD01001983 JAC86906.1 GBBI01001865 JAC16847.1 GECL01002811 JAP03313.1 GDHF01031817 JAI20497.1 HACA01033482 CDW50843.1 NIVC01001798 PAA63957.1 GL887696 EGI70382.1 HACA01031034 CDW48395.1 NIVC01001731 PAA64652.1 NIVC01000557 PAA80943.1 DQ174779 ABA55502.1 AM906135 CAP20049.1 AM231072 CAJ76986.1 AM231082 CAJ76996.1 AM231070 CAJ76984.1 AM231069 CAJ76983.1 AM231079 CAJ76993.1 AM231074 CAJ76988.1 AM231080 CAJ76994.1 AM906136 CAP20050.1 AJWK01006655 AM906142 AM906144 CAP20056.1 AM906145 CAP20059.1 AM906133 CAP20047.1 MTYJ01000027 OQV20875.1 AM906154 CAP20068.1 AM906153 CAP20067.1 AM906141 CAP20055.1 GECU01010428 JAS97278.1 GAKP01022166 GAKP01022163 JAC36789.1 U49974 AAC52011.1 MTYJ01000134 OQV12893.1 NSIT01000395 PJE77748.1 GGMS01009487 MBY78690.1 GGMS01017861 MBY87064.1 NEVH01006731 PNF36801.1 MTYJ01000064 OQV17200.1 GBBI01001742 JAC16970.1 KX930994 APL98287.1 ABLF02005939 LBMM01017269 KMQ84139.1 GBBI01001812 JAC16900.1 NEVH01020956 NEVH01016344 NEVH01006723 NEVH01003746 PNF20350.1 PNF25331.1 PNF37025.1 PNF40184.1 GEBQ01001581 JAT38396.1 GGMS01002163 MBY71366.1 KQ979265 KYN22061.1 MWRG01009041 PRD27309.1 NEVH01007822 PNF35315.1 GFTR01005237 JAW11189.1 NEVH01013196 PNF30090.1 NEVH01000595 PNF43644.1 GL438759 EFN68500.1 KX930999 APL98292.1 ABLF02008389 ABLF02015395 ABLF02015399 LBMM01010330 KMQ87532.1 HACA01020102 CDW37463.1 LBMM01024415 KMQ82581.1 KX930995 APL98288.1 GECU01035151 JAS72555.1 NEVH01026109 PNF14609.1 GECZ01025183 GECZ01019668 GECZ01003636 JAS44586.1 JAS50101.1 JAS66133.1
GAKP01015435 JAC43517.1 KX930998 APL98291.1 GBGD01001983 JAC86906.1 GBBI01001865 JAC16847.1 GECL01002811 JAP03313.1 GDHF01031817 JAI20497.1 HACA01033482 CDW50843.1 NIVC01001798 PAA63957.1 GL887696 EGI70382.1 HACA01031034 CDW48395.1 NIVC01001731 PAA64652.1 NIVC01000557 PAA80943.1 DQ174779 ABA55502.1 AM906135 CAP20049.1 AM231072 CAJ76986.1 AM231082 CAJ76996.1 AM231070 CAJ76984.1 AM231069 CAJ76983.1 AM231079 CAJ76993.1 AM231074 CAJ76988.1 AM231080 CAJ76994.1 AM906136 CAP20050.1 AJWK01006655 AM906142 AM906144 CAP20056.1 AM906145 CAP20059.1 AM906133 CAP20047.1 MTYJ01000027 OQV20875.1 AM906154 CAP20068.1 AM906153 CAP20067.1 AM906141 CAP20055.1 GECU01010428 JAS97278.1 GAKP01022166 GAKP01022163 JAC36789.1 U49974 AAC52011.1 MTYJ01000134 OQV12893.1 NSIT01000395 PJE77748.1 GGMS01009487 MBY78690.1 GGMS01017861 MBY87064.1 NEVH01006731 PNF36801.1 MTYJ01000064 OQV17200.1 GBBI01001742 JAC16970.1 KX930994 APL98287.1 ABLF02005939 LBMM01017269 KMQ84139.1 GBBI01001812 JAC16900.1 NEVH01020956 NEVH01016344 NEVH01006723 NEVH01003746 PNF20350.1 PNF25331.1 PNF37025.1 PNF40184.1 GEBQ01001581 JAT38396.1 GGMS01002163 MBY71366.1 KQ979265 KYN22061.1 MWRG01009041 PRD27309.1 NEVH01007822 PNF35315.1 GFTR01005237 JAW11189.1 NEVH01013196 PNF30090.1 NEVH01000595 PNF43644.1 GL438759 EFN68500.1 KX930999 APL98292.1 ABLF02008389 ABLF02015395 ABLF02015399 LBMM01010330 KMQ87532.1 HACA01020102 CDW37463.1 LBMM01024415 KMQ82581.1 KX930995 APL98288.1 GECU01035151 JAS72555.1 NEVH01026109 PNF14609.1 GECZ01025183 GECZ01019668 GECZ01003636 JAS44586.1 JAS50101.1 JAS66133.1
Proteomes
Interpro
SUPFAM
SSF53098
SSF53098
Gene 3D
ProteinModelPortal
Q1HPJ3
A0A0J7K8F0
A0A1B6G5C6
A0A034VN11
A0A1L5BY10
A0A069DT60
+ More
A0A023F6H6 A0A0V0G5J0 A0A0K8U1A4 A0A0K2VK65 A0A267ESW3 A0A1I8HNK7 F4W604 A0A0K2VDK7 A0A267EV83 A0A267G4K5 Q0QXC1 B3CK18 A6GV71 B7ZDI8 A6GV70 A6GV69 B7ZDI6 A6GV72 B7ZDI7 B3CK19 A0A1B0CCE3 B3CK24 B3CK26 B3CK17 A0A1W0X0B6 B3CK30 B3CK29 B3CK23 A0A1B6JDU2 A0A034V2R5 Q13539 A0A1W0WCL5 A0A2H9T3D9 A0A2S2QMC5 A0A2S2RAU4 A0A2J7R7K0 A0A1W0WPR3 A0A023F6W4 A0A1L5BY05 X1X2D9 A0A0J7K1H8 A0A023F6P2 A0A2J7Q9R7 A0A1B6MR58 A0A2S2Q0U2 A0A151JAL8 A0A2P6KLU2 A0A2J7R3C8 A0A224XTA3 A0A2J7QND6 A0A2J7RS37 E2ADI9 A0A1L5BY09 J9LNR4 A0A3Q2GQF1 A0A0J7KB73 A0A0K2UGY4 A0A0J7JXI1 A0A1L5BXZ8 A0A1B6HD45 A0A1I8HRU7 A0A2J7PE56 A0A1B6F2S5
A0A023F6H6 A0A0V0G5J0 A0A0K8U1A4 A0A0K2VK65 A0A267ESW3 A0A1I8HNK7 F4W604 A0A0K2VDK7 A0A267EV83 A0A267G4K5 Q0QXC1 B3CK18 A6GV71 B7ZDI8 A6GV70 A6GV69 B7ZDI6 A6GV72 B7ZDI7 B3CK19 A0A1B0CCE3 B3CK24 B3CK26 B3CK17 A0A1W0X0B6 B3CK30 B3CK29 B3CK23 A0A1B6JDU2 A0A034V2R5 Q13539 A0A1W0WCL5 A0A2H9T3D9 A0A2S2QMC5 A0A2S2RAU4 A0A2J7R7K0 A0A1W0WPR3 A0A023F6W4 A0A1L5BY05 X1X2D9 A0A0J7K1H8 A0A023F6P2 A0A2J7Q9R7 A0A1B6MR58 A0A2S2Q0U2 A0A151JAL8 A0A2P6KLU2 A0A2J7R3C8 A0A224XTA3 A0A2J7QND6 A0A2J7RS37 E2ADI9 A0A1L5BY09 J9LNR4 A0A3Q2GQF1 A0A0J7KB73 A0A0K2UGY4 A0A0J7JXI1 A0A1L5BXZ8 A0A1B6HD45 A0A1I8HRU7 A0A2J7PE56 A0A1B6F2S5
PDB
3F2K
E-value=1.03485e-14,
Score=189
Ontologies
Topology
Subcellular location
Cell membrane
Cell junction
Gap junction
Cell junction
Gap junction
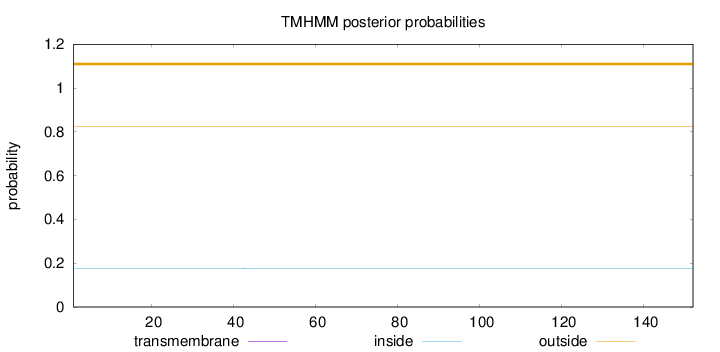
Length:
152
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00286
Exp number, first 60 AAs:
0.00286
Total prob of N-in:
0.17516
outside
1 - 152
Population Genetic Test Statistics
Pi
126.060407
Theta
85.551436
Tajima's D
1.201384
CLR
0
CSRT
0.714114294285286
Interpretation
Uncertain
