Gene
KWMTBOMO12442 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002355
Annotation
Bifunctional_methylenetetrahydrofolate_dehydrogenase/cyclohydrolase?_mitochondrial_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.945 Mitochondrial Reliability : 1.873
Sequence
CDS
ATGAGAAGTCACACATACATTGATTCCTTGGGCTCTGCCAGCATCATGGCGCGTATCCTCGATGGCAAAGCACTTGCTATGGAGATAAAGAAGGAGCTGAAGCAAAAAATAGCACAGTGGGTGAGTCTTGGAAACCGAGCGCCCACCATTCGCTGTATAATTGTTGGCGATGACCCAGCGAGCCACACCTATGTCAGGAACAAAGTTGAAGCCGCAAAATTTGTGGGCATCGATGCATTAACAATTAACAGGGACAGTGATATAACCGAAGAACAACTTCTCTCTGAGATCCAAAATTTAAATGAAGATAATAGTGTTGATGGAATTCTCGTCCAATTACCTGTACCTGATACAATTAATGAGAGGAGAGTTTGCGATGCTATTGCTCCTGAAAAAGATATCGATGGTTTCCACATAATCAACATCGGAAGGCTGTGTGTGGATCTACCTACCATTGTGCCGGCTACAGCATTGGCTGTAGTGGAAATGCTGAAAAGATTTAACATCGACACATTCGGTCGCAACGCGGTCGTGATAGGTCGTTCCAAAAACGTCGGTATGCCTATAGCTATGATGCTGCATAGCGACAAGAGACACGAAAGCGGATTAGGGATGGACGCTACCGTAACCATCTGCCATCGTTACACGCCAAAAGACAAACTGGAAGCATATTGTCGGAACGCTGATATCATTATCACCGCTACCGGTTTGCCCAAATTAATCAAAGCGGATATGGTGAAACCGGGCGCTACGATAATAGACGTTGGCATTACAAGAATAACCGATGAAAATGGAAAGTCTAGACTCGTCGGGGACGTTGATTACGAAGAAGTATCTAAGATCGCCGGTGCTATAACTCCAGTTCCCGGCGGAGTGGGTCCTATGACCGTGGCGATGCTGATGCATAACACATTCCAAGCAGCTCAGAGTCAAGCAAACAAATCCAATTAA
Protein
MRSHTYIDSLGSASIMARILDGKALAMEIKKELKQKIAQWVSLGNRAPTIRCIIVGDDPASHTYVRNKVEAAKFVGIDALTINRDSDITEEQLLSEIQNLNEDNSVDGILVQLPVPDTINERRVCDAIAPEKDIDGFHIINIGRLCVDLPTIVPATALAVVEMLKRFNIDTFGRNAVVIGRSKNVGMPIAMMLHSDKRHESGLGMDATVTICHRYTPKDKLEAYCRNADIIITATGLPKLIKADMVKPGATIIDVGITRITDENGKSRLVGDVDYEEVSKIAGAITPVPGGVGPMTVAMLMHNTFQAAQSQANKSN
Summary
Uniprot
H9IYM1
A0A2A4K8Z4
A0A3S2LQ25
A0A2H1VMW1
A0A194RGD9
A0A194Q023
+ More
A0A212F9K8 I4DRE1 A0A336MQG7 A0A336LQF1 T1DGQ4 A0A2M4BVC8 A0A2M4BVJ7 A0A2M3Z4G0 A0A182FLP3 A0A2M3Z4J1 T1E8H6 A0A2M4A235 A0A2M4CTT0 B0WDF0 A0A1Q3FHA6 A0A1Q3FHD2 A0A1A9Z6I6 A0A023ERE2 A0A1S4G338 A0A182ND91 Q16F77 A0A1B0GGJ8 D3TPH5 A0A182HEM5 A0A1A9V0D6 A7UTF7 A0A1I8MH51 A0A1I8MH50 A0A1B0CEH0 A0A182YPR9 A0A182WAY4 T1PNE5 A0A1I8MH59 A0A023EN06 A0A1A9YR99 W8BM65 A0A182HFB8 A0A182LLQ5 Q7PHT4 A0A084W180 A0A034VG54 A0A0C9PI39 A0A1L8DZN2 A0A034VKE6 A0A0A1XBW6 A0A182MBN9 A0A1I8PQC5 A0A1I8PQ71 A0A0K8TTX9 A0A023EP26 Q0IEF8 R7TMV7 A0A0K8W701 A0A0K8WL41 A0A0K8WEF6 D6WUR7 A0A091SSP4 B4NL93 N6TDW5 A0A182PVB4 W5N2G4 A0A2J7PDE7 A0A0T6AV37 A0A093P0G3 A0A151N579 A0A1B8Y8J3 A0A093QT07 A0A067RE48 A0A2P8ZNM3 A0A452HB80 R0K936 A0A093HWC7 A0A0L0CE32 A0A087R264 A0A1U8CVC2 A0A3B3QCB0 A0A151N5H6 A0A091PB02 A0A1S3HGZ1 A0A093EI54 A0A091L0W0 A0A3P8VWH8 A0A0A0AJM7 A0A1B6ETF3 A0A093QN91 A0A091QPN5 A0A093J3W0 A0A091WCL0 R7UT61 A0A091H216 A0A091T5X6 A0A093EY16 A0A091MN28 F6ZS18
A0A212F9K8 I4DRE1 A0A336MQG7 A0A336LQF1 T1DGQ4 A0A2M4BVC8 A0A2M4BVJ7 A0A2M3Z4G0 A0A182FLP3 A0A2M3Z4J1 T1E8H6 A0A2M4A235 A0A2M4CTT0 B0WDF0 A0A1Q3FHA6 A0A1Q3FHD2 A0A1A9Z6I6 A0A023ERE2 A0A1S4G338 A0A182ND91 Q16F77 A0A1B0GGJ8 D3TPH5 A0A182HEM5 A0A1A9V0D6 A7UTF7 A0A1I8MH51 A0A1I8MH50 A0A1B0CEH0 A0A182YPR9 A0A182WAY4 T1PNE5 A0A1I8MH59 A0A023EN06 A0A1A9YR99 W8BM65 A0A182HFB8 A0A182LLQ5 Q7PHT4 A0A084W180 A0A034VG54 A0A0C9PI39 A0A1L8DZN2 A0A034VKE6 A0A0A1XBW6 A0A182MBN9 A0A1I8PQC5 A0A1I8PQ71 A0A0K8TTX9 A0A023EP26 Q0IEF8 R7TMV7 A0A0K8W701 A0A0K8WL41 A0A0K8WEF6 D6WUR7 A0A091SSP4 B4NL93 N6TDW5 A0A182PVB4 W5N2G4 A0A2J7PDE7 A0A0T6AV37 A0A093P0G3 A0A151N579 A0A1B8Y8J3 A0A093QT07 A0A067RE48 A0A2P8ZNM3 A0A452HB80 R0K936 A0A093HWC7 A0A0L0CE32 A0A087R264 A0A1U8CVC2 A0A3B3QCB0 A0A151N5H6 A0A091PB02 A0A1S3HGZ1 A0A093EI54 A0A091L0W0 A0A3P8VWH8 A0A0A0AJM7 A0A1B6ETF3 A0A093QN91 A0A091QPN5 A0A093J3W0 A0A091WCL0 R7UT61 A0A091H216 A0A091T5X6 A0A093EY16 A0A091MN28 F6ZS18
Pubmed
19121390
26354079
22118469
22651552
24330624
24945155
+ More
17510324 20353571 26483478 12364791 14747013 17210077 25315136 25244985 24495485 20966253 24438588 25348373 25830018 26369729 23254933 18362917 19820115 17994087 23537049 22293439 20431018 24845553 29403074 28562605 26108605 29240929 24487278
17510324 20353571 26483478 12364791 14747013 17210077 25315136 25244985 24495485 20966253 24438588 25348373 25830018 26369729 23254933 18362917 19820115 17994087 23537049 22293439 20431018 24845553 29403074 28562605 26108605 29240929 24487278
EMBL
BABH01037918
LC366023
BBC53721.1
NWSH01000051
PCG80180.1
RSAL01000033
+ More
RVE51492.1 ODYU01003424 SOQ42169.1 KQ460211 KPJ16514.1 KQ459585 KPI98344.1 AGBW02009565 OWR50410.1 AK405080 BAM20481.1 UFQT01001832 SSX31925.1 UFQT01000115 SSX20276.1 GALA01001571 JAA93281.1 GGFJ01007811 MBW56952.1 GGFJ01007810 MBW56951.1 GGFM01002656 MBW23407.1 GGFM01002664 MBW23415.1 GAMD01002172 JAA99418.1 GGFK01001494 MBW34815.1 GGFL01004576 MBW68754.1 DS231897 EDS44521.1 GFDL01008108 JAV26937.1 GFDL01008106 JAV26939.1 GAPW01002377 JAC11221.1 CH478498 EAT32889.1 CCAG010006689 EZ423327 ADD19603.1 JXUM01035647 JXUM01151840 KQ571311 KQ561068 KXJ68206.1 KXJ79808.1 AAAB01008942 EDO64016.1 AJWK01008866 KA649595 AFP64224.1 GAPW01002830 JAC10768.1 GAMC01004365 JAC02191.1 JXUM01036772 JXUM01036773 JXUM01036774 KQ561105 KXJ79689.1 EAA44412.3 ATLV01019244 KE525266 KFB43974.1 GAKP01016681 JAC42271.1 GBYB01000568 JAG70335.1 GFDF01002195 JAV11889.1 GAKP01016682 JAC42270.1 GBXI01006339 JAD07953.1 AXCM01010523 GDAI01000193 JAI17410.1 GAPW01002883 JAC10715.1 CH477691 EAT37237.1 AMQN01011953 AMQN01011954 KB309204 ELT95208.1 GDHF01005495 JAI46819.1 GDHF01000522 JAI51792.1 GDHF01002890 JAI49424.1 KQ971363 EFA07809.2 KK485136 KFQ61742.1 CH964272 EDW84296.2 APGK01041496 APGK01041497 KB740994 KB632356 ENN75943.1 ERL93399.1 AHAT01017245 NEVH01026389 PNF14360.1 LJIG01022796 KRT78670.1 KL225121 KFW67680.1 AKHW03004011 KYO31958.1 KV460381 OCA19320.1 KL431109 KFW91651.1 KK852562 KDR21308.1 PYGN01000008 PSN58111.1 KB742619 EOB06322.1 KL206808 KFV85931.1 JRES01000519 KNC30457.1 KL226056 KFM07568.1 KYO31957.1 KK670420 KFQ04556.1 KL244970 KFV14216.1 KK764453 KFP45922.1 KL872182 KGL94709.1 GECZ01028527 GECZ01019906 JAS41242.1 JAS49863.1 KL672314 KFW85447.1 KK800824 KFQ28591.1 KK601672 KFW06552.1 KK735164 KFR12860.1 AMQN01007383 AMQN01007384 KB300556 ELU06551.1 KL521750 KFO89876.1 KK437751 KFQ69245.1 KK612139 KFV46759.1 KK833161 KFP78075.1 AAMC01118666 AAMC01118667 AAMC01118668 AAMC01118669
RVE51492.1 ODYU01003424 SOQ42169.1 KQ460211 KPJ16514.1 KQ459585 KPI98344.1 AGBW02009565 OWR50410.1 AK405080 BAM20481.1 UFQT01001832 SSX31925.1 UFQT01000115 SSX20276.1 GALA01001571 JAA93281.1 GGFJ01007811 MBW56952.1 GGFJ01007810 MBW56951.1 GGFM01002656 MBW23407.1 GGFM01002664 MBW23415.1 GAMD01002172 JAA99418.1 GGFK01001494 MBW34815.1 GGFL01004576 MBW68754.1 DS231897 EDS44521.1 GFDL01008108 JAV26937.1 GFDL01008106 JAV26939.1 GAPW01002377 JAC11221.1 CH478498 EAT32889.1 CCAG010006689 EZ423327 ADD19603.1 JXUM01035647 JXUM01151840 KQ571311 KQ561068 KXJ68206.1 KXJ79808.1 AAAB01008942 EDO64016.1 AJWK01008866 KA649595 AFP64224.1 GAPW01002830 JAC10768.1 GAMC01004365 JAC02191.1 JXUM01036772 JXUM01036773 JXUM01036774 KQ561105 KXJ79689.1 EAA44412.3 ATLV01019244 KE525266 KFB43974.1 GAKP01016681 JAC42271.1 GBYB01000568 JAG70335.1 GFDF01002195 JAV11889.1 GAKP01016682 JAC42270.1 GBXI01006339 JAD07953.1 AXCM01010523 GDAI01000193 JAI17410.1 GAPW01002883 JAC10715.1 CH477691 EAT37237.1 AMQN01011953 AMQN01011954 KB309204 ELT95208.1 GDHF01005495 JAI46819.1 GDHF01000522 JAI51792.1 GDHF01002890 JAI49424.1 KQ971363 EFA07809.2 KK485136 KFQ61742.1 CH964272 EDW84296.2 APGK01041496 APGK01041497 KB740994 KB632356 ENN75943.1 ERL93399.1 AHAT01017245 NEVH01026389 PNF14360.1 LJIG01022796 KRT78670.1 KL225121 KFW67680.1 AKHW03004011 KYO31958.1 KV460381 OCA19320.1 KL431109 KFW91651.1 KK852562 KDR21308.1 PYGN01000008 PSN58111.1 KB742619 EOB06322.1 KL206808 KFV85931.1 JRES01000519 KNC30457.1 KL226056 KFM07568.1 KYO31957.1 KK670420 KFQ04556.1 KL244970 KFV14216.1 KK764453 KFP45922.1 KL872182 KGL94709.1 GECZ01028527 GECZ01019906 JAS41242.1 JAS49863.1 KL672314 KFW85447.1 KK800824 KFQ28591.1 KK601672 KFW06552.1 KK735164 KFR12860.1 AMQN01007383 AMQN01007384 KB300556 ELU06551.1 KL521750 KFO89876.1 KK437751 KFQ69245.1 KK612139 KFV46759.1 KK833161 KFP78075.1 AAMC01118666 AAMC01118667 AAMC01118668 AAMC01118669
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000069272 UP000002320 UP000092445 UP000075884 UP000008820 UP000092444 UP000069940 UP000249989 UP000078200 UP000007062 UP000095301 UP000092461 UP000076408 UP000075920 UP000092443 UP000075882 UP000030765 UP000075883 UP000095300 UP000014760 UP000007266 UP000007798 UP000019118 UP000030742 UP000075885 UP000018468 UP000235965 UP000054081 UP000050525 UP000008143 UP000027135 UP000245037 UP000291020 UP000053584 UP000037069 UP000053286 UP000189705 UP000261540 UP000085678 UP000265120 UP000053858 UP000053258 UP000053605
UP000069272 UP000002320 UP000092445 UP000075884 UP000008820 UP000092444 UP000069940 UP000249989 UP000078200 UP000007062 UP000095301 UP000092461 UP000076408 UP000075920 UP000092443 UP000075882 UP000030765 UP000075883 UP000095300 UP000014760 UP000007266 UP000007798 UP000019118 UP000030742 UP000075885 UP000018468 UP000235965 UP000054081 UP000050525 UP000008143 UP000027135 UP000245037 UP000291020 UP000053584 UP000037069 UP000053286 UP000189705 UP000261540 UP000085678 UP000265120 UP000053858 UP000053258 UP000053605
PRIDE
Pfam
Interpro
IPR000672
THF_DH/CycHdrlase
+ More
IPR020630 THF_DH/CycHdrlase_cat_dom
IPR036291 NAD(P)-bd_dom_sf
IPR020631 THF_DH/CycHdrlase_NAD-bd_dom
IPR020867 THF_DH/CycHdrlase_CS
IPR006612 THAP_Znf
IPR008967 p53-like_TF_DNA-bd
IPR002110 Ankyrin_rpt
IPR033926 IPT_NFkappaB
IPR013783 Ig-like_fold
IPR011539 RHD_DNA_bind_dom
IPR000451 NFkB/Dor
IPR014756 Ig_E-set
IPR002909 IPT_dom
IPR032397 RHD_dimer
IPR036770 Ankyrin_rpt-contain_sf
IPR037059 RHD_DNA_bind_dom_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR020630 THF_DH/CycHdrlase_cat_dom
IPR036291 NAD(P)-bd_dom_sf
IPR020631 THF_DH/CycHdrlase_NAD-bd_dom
IPR020867 THF_DH/CycHdrlase_CS
IPR006612 THAP_Znf
IPR008967 p53-like_TF_DNA-bd
IPR002110 Ankyrin_rpt
IPR033926 IPT_NFkappaB
IPR013783 Ig-like_fold
IPR011539 RHD_DNA_bind_dom
IPR000451 NFkB/Dor
IPR014756 Ig_E-set
IPR002909 IPT_dom
IPR032397 RHD_dimer
IPR036770 Ankyrin_rpt-contain_sf
IPR037059 RHD_DNA_bind_dom_sf
IPR020683 Ankyrin_rpt-contain_dom
Gene 3D
ProteinModelPortal
H9IYM1
A0A2A4K8Z4
A0A3S2LQ25
A0A2H1VMW1
A0A194RGD9
A0A194Q023
+ More
A0A212F9K8 I4DRE1 A0A336MQG7 A0A336LQF1 T1DGQ4 A0A2M4BVC8 A0A2M4BVJ7 A0A2M3Z4G0 A0A182FLP3 A0A2M3Z4J1 T1E8H6 A0A2M4A235 A0A2M4CTT0 B0WDF0 A0A1Q3FHA6 A0A1Q3FHD2 A0A1A9Z6I6 A0A023ERE2 A0A1S4G338 A0A182ND91 Q16F77 A0A1B0GGJ8 D3TPH5 A0A182HEM5 A0A1A9V0D6 A7UTF7 A0A1I8MH51 A0A1I8MH50 A0A1B0CEH0 A0A182YPR9 A0A182WAY4 T1PNE5 A0A1I8MH59 A0A023EN06 A0A1A9YR99 W8BM65 A0A182HFB8 A0A182LLQ5 Q7PHT4 A0A084W180 A0A034VG54 A0A0C9PI39 A0A1L8DZN2 A0A034VKE6 A0A0A1XBW6 A0A182MBN9 A0A1I8PQC5 A0A1I8PQ71 A0A0K8TTX9 A0A023EP26 Q0IEF8 R7TMV7 A0A0K8W701 A0A0K8WL41 A0A0K8WEF6 D6WUR7 A0A091SSP4 B4NL93 N6TDW5 A0A182PVB4 W5N2G4 A0A2J7PDE7 A0A0T6AV37 A0A093P0G3 A0A151N579 A0A1B8Y8J3 A0A093QT07 A0A067RE48 A0A2P8ZNM3 A0A452HB80 R0K936 A0A093HWC7 A0A0L0CE32 A0A087R264 A0A1U8CVC2 A0A3B3QCB0 A0A151N5H6 A0A091PB02 A0A1S3HGZ1 A0A093EI54 A0A091L0W0 A0A3P8VWH8 A0A0A0AJM7 A0A1B6ETF3 A0A093QN91 A0A091QPN5 A0A093J3W0 A0A091WCL0 R7UT61 A0A091H216 A0A091T5X6 A0A093EY16 A0A091MN28 F6ZS18
A0A212F9K8 I4DRE1 A0A336MQG7 A0A336LQF1 T1DGQ4 A0A2M4BVC8 A0A2M4BVJ7 A0A2M3Z4G0 A0A182FLP3 A0A2M3Z4J1 T1E8H6 A0A2M4A235 A0A2M4CTT0 B0WDF0 A0A1Q3FHA6 A0A1Q3FHD2 A0A1A9Z6I6 A0A023ERE2 A0A1S4G338 A0A182ND91 Q16F77 A0A1B0GGJ8 D3TPH5 A0A182HEM5 A0A1A9V0D6 A7UTF7 A0A1I8MH51 A0A1I8MH50 A0A1B0CEH0 A0A182YPR9 A0A182WAY4 T1PNE5 A0A1I8MH59 A0A023EN06 A0A1A9YR99 W8BM65 A0A182HFB8 A0A182LLQ5 Q7PHT4 A0A084W180 A0A034VG54 A0A0C9PI39 A0A1L8DZN2 A0A034VKE6 A0A0A1XBW6 A0A182MBN9 A0A1I8PQC5 A0A1I8PQ71 A0A0K8TTX9 A0A023EP26 Q0IEF8 R7TMV7 A0A0K8W701 A0A0K8WL41 A0A0K8WEF6 D6WUR7 A0A091SSP4 B4NL93 N6TDW5 A0A182PVB4 W5N2G4 A0A2J7PDE7 A0A0T6AV37 A0A093P0G3 A0A151N579 A0A1B8Y8J3 A0A093QT07 A0A067RE48 A0A2P8ZNM3 A0A452HB80 R0K936 A0A093HWC7 A0A0L0CE32 A0A087R264 A0A1U8CVC2 A0A3B3QCB0 A0A151N5H6 A0A091PB02 A0A1S3HGZ1 A0A093EI54 A0A091L0W0 A0A3P8VWH8 A0A0A0AJM7 A0A1B6ETF3 A0A093QN91 A0A091QPN5 A0A093J3W0 A0A091WCL0 R7UT61 A0A091H216 A0A091T5X6 A0A093EY16 A0A091MN28 F6ZS18
PDB
5ZF1
E-value=1.64178e-165,
Score=1494
Ontologies
PATHWAY
GO
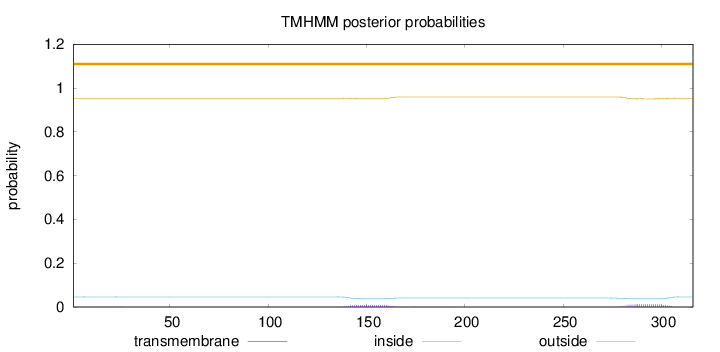
Topology
Length:
316
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.46581
Exp number, first 60 AAs:
0.00550999999999999
Total prob of N-in:
0.04615
outside
1 - 316
Population Genetic Test Statistics
Pi
152.386063
Theta
208.960765
Tajima's D
-1.286334
CLR
259.5045
CSRT
0.0890455477226139
Interpretation
Uncertain
