Gene
KWMTBOMO12441
Pre Gene Modal
BGIBMGA002354
Annotation
Furin-like_protease_2_[Papilio_machaon]
Full name
Furin-like protease 2
Location in the cell
Extracellular Reliability : 3.664
Sequence
CDS
ATGAGGCAATCTAGCGCACCGTCTTTTCCTGATCCACTGTTCAAAGAGCAGTGGTATTTGAATGGTGGCGCAGCTGATGGTCTCGATATGAATGTCGGGTATGCTTGGAGGAAGGGCTATACGGGGAAAGGTGTCGTCATTACTATACTCGATGATGGCATACAGCCTAATCACCCGGATCTATCACAGAATTACGATCCGGCCGCTTCAACTGACATAAACGGGAACGATACGGACCCGACACCACAGGACAATGGCGACAATAAACACGGGACCCGTTGTGCAGGAGAGGTAGCTGCCGTCGCTTATAACCGATACTGTGGCGTTGGTATAGCATACAATGCTAGCATTGGAGGAGTGAGAATGCTGGATGGCGTTGTAAACGACGCAGTGGAAGCTCAAGCACTCGGATTTAACACGCATCATATTGATATATATAGTGCGTCGTGGGGACCGGAGGACGACGGAAAAACTGTCGACGGCCCGGGTCCTTTAGCTCGAAGAGCGTTTATCAACGGAGTAACGAACGGAAGAGGGGGAAAAGGATCCATATTCATATGGGCATCAGGAAACGGGGGGCGGCACACTGACTCTTGTAACTGCGACGGGTATGCTAATAGCATATTCACTATATCCATATCTAGCGCAACGCAAGGAGGGTACAAGCCTTGGTATTTAGAAGAATGCTCTTCGACTTTAGCTTCAACGTATAGTTCAGGTACACCAGGACGAGATAAAAGCGTTGCAACCGTTGATATGGACGTCCAATTACGAGCGGAGCATATCTGCACGGTCGACCATACGGGTACCTCAGCATCCGCACCATTGGCTGCAGGAATTTGTGCATTAGCTTTAGAAGCTAATCCGCTATTGTCGTGGAGAGATATGCAGCATTTAATTGTAATGACGTCACGTTCTCAACCCCTAGAAAAAGAAGAAGGGTGGATCGTGAATGGTGTTAAAAGAAAAGTTAGTCATAAATTCGGTTACGGACTGATGGATGCAGCTCAAATGGTCAATCTAGCTGAGCAGTGGACTAATGTGGCGCCTCAGCACATTTGTAAATCCCAAGAAATAAACGAAGACAAATCCATTGAAACGTCTTTCGGTTATACGATATCTGTACACATGGACGTGAATGGGTGCAGCGGGACGACAAACGAGGTCAGATTTTTGGAACACGTTCAATGTAAAATTTCGTTAAGCTTTTTTCCGAGAGGTAATTTACGAATATTGCTTACATCACCGATGGGAACCACATCTACGCTTCTATTCGAAAGGACCCATGACGCAATTAGTTCGAATTTCGACGACTGGCCGTTTTTGAGCGTTCATTTCTGGGGTGAAAACGCCGAAGGGAGATGGACTTTGCAAATTATAAATGCGGGCAATACTCACGCGACGCAAGCTGGAGTATTGAAGAAATGGCAATTATTTTTTTACGGTACCGCTGTCTCCCCCATACGGCTACGAAATAAAGACTATATGAACTCGAAAAATTCGTACCACGAAGAAAAAACATATCACATTAATGACGTATACGATGCATCTGAATACTCTCAGTTTTTAAACGAAATTGAACTAGGTATTTCAGACAGGCGTAGCTACGAGAAAAACATTCCATCGGCTCAAAGAAAGAATGTTTTAGCGGATGCTAATGATAAGCAAGTGCAGAGACTATGCGATCCGGAATGTGATTCGCAGGGCTGCTACGGCAAAGGCCCAACGCAGTGTGTCGCTTGTAAACATTACCGTTTGGATAATTCATGTGTTTCAAAATGCCCCCCCAGGAGTTTTGTTAATCAAGGCGGAGTCTGCTGGCCATGTCACGAGTCTTGTGAAACGTGTGCCGGAGCCGGACAAGATTCATGTTTGACTTGCGCGCCGGCACATTTACTGGTCGTGGATTTGGCTGTCTGCCTGCAGCAGTGCCCAGATGGTTACTATGAAGACCCAGATGCCAACGCATGTTTTCCATGCGCCGAACATTGTGATACATGCACAGAGAAAGCAGACTTGTGTTCTTCTTGTGCCCACAGTTATGTTTTGTACAATGGATCTTGTTTAGCGGCCTGTCCACCAAGTACTTATCAAAAAGACTACTATGGCTGTATGCCATGTCACGAATCTTGTGACTCTTGTTACGGACCGAGCCAAAATTCTTGCGTGTCCTGTAAAATTGGAGACTTTGTCTTCAAGAACCAATGTATTTCGAAGTGTCCTTCAGGCTTCTATGCGGATTCGCAGAAAAGGGAATGCTTAGAGTGTCCGATTGGATGTTTGGTATGCTCTTATGGAGTGTGTTCTTCCTGTAAAGAGGAGTGGACGCTCACTAAGGGAGGAAGTTGCTTTCCCGATGGAAACGAAAAATGTGATACAAATGAATATTACGAGGGAGGTCGATGCAAAAATTGCCATTCAACATGCGAGAAATGTAGCGGACCGAACGAGTGGGATTGCATCTCTTGTTCAAGTCCTCTTCTGTTACAAGGGTCGAGGTGTGTGGCAGAATGTGGACGCGGGTATTATCAAACCGCCGGGTCTGGACCAACAAGTTGTGTTACTTGCGCTCATCCACTTCAATTAGATAGAGTCAACTACAAATGTTTACCTTGCTGCTTAGAAAATATGGCATCTATTTACTTAACAGTCAATGAAACATCAGATTGTTGCCATTGTGATAAGGATATCGGGGGGTGCTTAAATGGTTCATCGGCTGGTAAAAGAAGAATAGCTGAAAATATAAGGACACATATTACTGCATCATATTTCATAGAGGATTCGAAGGATTCTGCAGATACCATCGATTTAAACTATGTTATCATTTGGAGTGCGGCAGCTACTGTTATTGTGATTGTCATAATCGACAGAAATTTATCTGATATTTCATAA
Protein
MRQSSAPSFPDPLFKEQWYLNGGAADGLDMNVGYAWRKGYTGKGVVITILDDGIQPNHPDLSQNYDPAASTDINGNDTDPTPQDNGDNKHGTRCAGEVAAVAYNRYCGVGIAYNASIGGVRMLDGVVNDAVEAQALGFNTHHIDIYSASWGPEDDGKTVDGPGPLARRAFINGVTNGRGGKGSIFIWASGNGGRHTDSCNCDGYANSIFTISISSATQGGYKPWYLEECSSTLASTYSSGTPGRDKSVATVDMDVQLRAEHICTVDHTGTSASAPLAAGICALALEANPLLSWRDMQHLIVMTSRSQPLEKEEGWIVNGVKRKVSHKFGYGLMDAAQMVNLAEQWTNVAPQHICKSQEINEDKSIETSFGYTISVHMDVNGCSGTTNEVRFLEHVQCKISLSFFPRGNLRILLTSPMGTTSTLLFERTHDAISSNFDDWPFLSVHFWGENAEGRWTLQIINAGNTHATQAGVLKKWQLFFYGTAVSPIRLRNKDYMNSKNSYHEEKTYHINDVYDASEYSQFLNEIELGISDRRSYEKNIPSAQRKNVLADANDKQVQRLCDPECDSQGCYGKGPTQCVACKHYRLDNSCVSKCPPRSFVNQGGVCWPCHESCETCAGAGQDSCLTCAPAHLLVVDLAVCLQQCPDGYYEDPDANACFPCAEHCDTCTEKADLCSSCAHSYVLYNGSCLAACPPSTYQKDYYGCMPCHESCDSCYGPSQNSCVSCKIGDFVFKNQCISKCPSGFYADSQKRECLECPIGCLVCSYGVCSSCKEEWTLTKGGSCFPDGNEKCDTNEYYEGGRCKNCHSTCEKCSGPNEWDCISCSSPLLLQGSRCVAECGRGYYQTAGSGPTSCVTCAHPLQLDRVNYKCLPCCLENMASIYLTVNETSDCCHCDKDIGGCLNGSSAGKRRIAENIRTHITASYFIEDSKDSADTIDLNYVIIWSAAATVIVIVIIDRNLSDIS
Summary
Description
Furin is likely to represent the ubiquitous endoprotease activity within constitutive secretory pathways and capable of cleavage at the RX(K/R)R consensus motif.
Catalytic Activity
Release of mature proteins from their proproteins by cleavage of -Arg-Xaa-Yaa-Arg-|-Zaa- bonds, where Xaa can be any amino acid and Yaa is Arg or Lys. Releases albumin, complement component C3 and von Willebrand factor from their respective precursors.
Cofactor
Ca(2+)
Similarity
Belongs to the peptidase S8 family.
Belongs to the peptidase S8 family. Furin subfamily.
Belongs to the peptidase S8 family. Furin subfamily.
Keywords
Alternative splicing
Cleavage on pair of basic residues
Complete proteome
Disulfide bond
Glycoprotein
Hydrolase
Membrane
Protease
Reference proteome
Repeat
Serine protease
Signal
Transmembrane
Transmembrane helix
Zymogen
Feature
chain Furin-like protease 2
splice variant In isoform A.
splice variant In isoform A.
Uniprot
H9IYM0
A0A194RGR3
A0A2A4JGI8
A0A2H1WK85
A0A437B8F0
A0A212FLJ6
+ More
A0A194PZ75 A0A0L7LPQ1 A0A2A4K7Y7 A0A212F4A7 Q26489 A0A437BM12 A0A194RFY5 A0A194PYD1 A0A2H1W496 H9IYN1 A0A0A1XGS4 A0A0K8U8C6 A0A0A1X1M3 A0A0L7LUE0 W8APW3 A0A1Q3FTU0 A0A1Q3FU12 A0A1Q3FTV5 A0A1Q3FUD1 T1P9R1 A0A1I8Q1H9 A0A1I8Q1I4 A0A0L0BQD1 A0A2M4DS50 A0A2M3Z0N1 A0A2M4A589 A0A2M4DS69 A0A2M4A5Q5 A0A2M4A4R7 A0A2M4B9F8 A0A2M4B9B2 A0A2M4CJY4 A0A2M4B9D5 Q5DNW1 A0A023EXA8 W4VRF9 A0A336LJ86 A0A336LIK8 A0A2C9GWR4 A0A195FL77 A0A1B6GHW2 A0A195BWS1 A0A1B6CCM9 A0A158NEY8 A0A1Y1KM81 A0A139WNE9 A0A1J1IF38 A0A0C9QJY7 A0A151IEG1 A0A0C9QQN8 A0A088AL19 A0A336LJV0 E2BLN8 A0A0Q9WMJ9 B4M1S6 A0A3B0K7Y0 A0A0R1EG80 B4L8R4 A0A0R3P5B9 B4PXS5 B3NX47 Q29GI8 A0A0Q5T754 A0A0M4ELX6 A0A2D3FB63 A0A0R1EEA9 A0A0Q9XGD4 B4NCL2 A0A0R3P6H0 E0V9N9 A0A0R1EGT9 A0A0Q9XJA6 E1JJL4 A0A0R3P116 A0A0Q9XRF1 P30432 A0A0Q5T532 B4JKM2 B3N0S2 P30432-2 B4R5T4 A0A0P8ZKG0 F4WVD8 A0A1S4GGJ0 A0A195EN83 A0A2A3EGB8 A0A151XIV9 A0A2H8TUQ2 A0A026X0H0 A0A1W4VTY2 A0A1W4VHD4 A0A084VGH8 X1WJ00 A0A1W4VVH0
A0A194PZ75 A0A0L7LPQ1 A0A2A4K7Y7 A0A212F4A7 Q26489 A0A437BM12 A0A194RFY5 A0A194PYD1 A0A2H1W496 H9IYN1 A0A0A1XGS4 A0A0K8U8C6 A0A0A1X1M3 A0A0L7LUE0 W8APW3 A0A1Q3FTU0 A0A1Q3FU12 A0A1Q3FTV5 A0A1Q3FUD1 T1P9R1 A0A1I8Q1H9 A0A1I8Q1I4 A0A0L0BQD1 A0A2M4DS50 A0A2M3Z0N1 A0A2M4A589 A0A2M4DS69 A0A2M4A5Q5 A0A2M4A4R7 A0A2M4B9F8 A0A2M4B9B2 A0A2M4CJY4 A0A2M4B9D5 Q5DNW1 A0A023EXA8 W4VRF9 A0A336LJ86 A0A336LIK8 A0A2C9GWR4 A0A195FL77 A0A1B6GHW2 A0A195BWS1 A0A1B6CCM9 A0A158NEY8 A0A1Y1KM81 A0A139WNE9 A0A1J1IF38 A0A0C9QJY7 A0A151IEG1 A0A0C9QQN8 A0A088AL19 A0A336LJV0 E2BLN8 A0A0Q9WMJ9 B4M1S6 A0A3B0K7Y0 A0A0R1EG80 B4L8R4 A0A0R3P5B9 B4PXS5 B3NX47 Q29GI8 A0A0Q5T754 A0A0M4ELX6 A0A2D3FB63 A0A0R1EEA9 A0A0Q9XGD4 B4NCL2 A0A0R3P6H0 E0V9N9 A0A0R1EGT9 A0A0Q9XJA6 E1JJL4 A0A0R3P116 A0A0Q9XRF1 P30432 A0A0Q5T532 B4JKM2 B3N0S2 P30432-2 B4R5T4 A0A0P8ZKG0 F4WVD8 A0A1S4GGJ0 A0A195EN83 A0A2A3EGB8 A0A151XIV9 A0A2H8TUQ2 A0A026X0H0 A0A1W4VTY2 A0A1W4VHD4 A0A084VGH8 X1WJ00 A0A1W4VVH0
EC Number
3.4.21.75
Pubmed
19121390
26354079
22118469
26227816
25830018
24495485
+ More
26108605 24945155 21347285 28004739 18362917 19820115 20798317 17994087 18057021 17550304 15632085 23185243 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1512259 7880443 12537569 21719571 12364791 24508170 24438588
26108605 24945155 21347285 28004739 18362917 19820115 20798317 17994087 18057021 17550304 15632085 23185243 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1512259 7880443 12537569 21719571 12364791 24508170 24438588
EMBL
BABH01037938
KQ460211
KPJ16515.1
NWSH01001572
PCG70828.1
ODYU01009227
+ More
SOQ53495.1 RSAL01000126 RVE46569.1 AGBW02007756 OWR54611.1 KQ459585 KPI98343.1 JTDY01000441 KOB77156.1 NWSH01000051 PCG80179.1 AGBW02010411 OWR48567.1 Z68888 CAA93116.1 RSAL01000033 RVE51494.1 KPJ16512.1 KPI98346.1 ODYU01006244 SOQ47911.1 BABH01037900 GBXI01004172 JAD10120.1 GDHF01029508 JAI22806.1 GBXI01009632 JAD04660.1 JTDY01000105 KOB78841.1 GAMC01018483 JAB88072.1 GFDL01004014 JAV31031.1 GFDL01004017 JAV31028.1 GFDL01004015 JAV31030.1 GFDL01004012 JAV31033.1 KA645434 AFP60063.1 JRES01001522 KNC22262.1 GGFL01016196 MBW80374.1 GGFM01001305 MBW22056.1 GGFK01002655 MBW35976.1 GGFL01016237 MBW80415.1 GGFK01002802 MBW36123.1 GGFK01002391 MBW35712.1 GGFJ01000501 MBW49642.1 GGFJ01000503 MBW49644.1 GGFL01001478 MBW65656.1 GGFJ01000508 MBW49649.1 AY555267 AAT37510.1 GAPW01000047 JAC13551.1 GANO01003419 JAB56452.1 UFQT01000021 SSX17946.1 SSX17944.1 KQ981490 KYN41168.1 GECZ01007763 JAS62006.1 KQ976401 KYM92418.1 GEDC01026106 JAS11192.1 ADTU01013412 ADTU01013413 GEZM01082529 JAV61340.1 KQ971311 KYB29424.1 CVRI01000047 CRK97614.1 GBYB01003844 JAG73611.1 KQ977871 KYM99195.1 GBYB01002977 JAG72744.1 SSX17945.1 GL449036 EFN83372.1 CH940651 KRF82319.1 EDW65630.1 KRF82320.1 OUUW01000015 SPP88772.1 CM000162 KRK06433.1 CH933815 EDW08039.1 CH379064 KRT06943.1 EDX01911.1 CH954180 EDV46876.2 EAL32121.2 KQS30224.1 KQS30225.1 CP012528 ALC48584.1 KY407546 ATU47252.1 KRK06435.1 KRG07562.1 KRG07564.1 KRG07565.1 KRG07567.1 CH964239 EDW82571.2 KRT06944.1 KRT06945.1 DS234995 EEB10108.1 KRK06434.1 KRK06436.1 KRG07561.1 AE014298 ACZ95309.1 KRT06942.1 KRG07566.1 M94375 L33831 BT021414 AY070553 KQS30223.1 CH916370 EDW00125.1 CH902644 EDV34801.1 CM000366 EDX18084.1 KPU75290.1 GL888387 EGI61759.1 AAAB01008859 KQ978625 KYN29621.1 KZ288254 PBC30823.1 KQ982080 KYQ60346.1 GFXV01006180 MBW17985.1 KK107064 EZA60894.1 ATLV01012931 ATLV01012932 ATLV01012933 ATLV01012934 ATLV01012935 ATLV01012936 ATLV01012937 ATLV01012938 KE524827 KFB37072.1 ABLF02040171 ABLF02040188 ABLF02040191
SOQ53495.1 RSAL01000126 RVE46569.1 AGBW02007756 OWR54611.1 KQ459585 KPI98343.1 JTDY01000441 KOB77156.1 NWSH01000051 PCG80179.1 AGBW02010411 OWR48567.1 Z68888 CAA93116.1 RSAL01000033 RVE51494.1 KPJ16512.1 KPI98346.1 ODYU01006244 SOQ47911.1 BABH01037900 GBXI01004172 JAD10120.1 GDHF01029508 JAI22806.1 GBXI01009632 JAD04660.1 JTDY01000105 KOB78841.1 GAMC01018483 JAB88072.1 GFDL01004014 JAV31031.1 GFDL01004017 JAV31028.1 GFDL01004015 JAV31030.1 GFDL01004012 JAV31033.1 KA645434 AFP60063.1 JRES01001522 KNC22262.1 GGFL01016196 MBW80374.1 GGFM01001305 MBW22056.1 GGFK01002655 MBW35976.1 GGFL01016237 MBW80415.1 GGFK01002802 MBW36123.1 GGFK01002391 MBW35712.1 GGFJ01000501 MBW49642.1 GGFJ01000503 MBW49644.1 GGFL01001478 MBW65656.1 GGFJ01000508 MBW49649.1 AY555267 AAT37510.1 GAPW01000047 JAC13551.1 GANO01003419 JAB56452.1 UFQT01000021 SSX17946.1 SSX17944.1 KQ981490 KYN41168.1 GECZ01007763 JAS62006.1 KQ976401 KYM92418.1 GEDC01026106 JAS11192.1 ADTU01013412 ADTU01013413 GEZM01082529 JAV61340.1 KQ971311 KYB29424.1 CVRI01000047 CRK97614.1 GBYB01003844 JAG73611.1 KQ977871 KYM99195.1 GBYB01002977 JAG72744.1 SSX17945.1 GL449036 EFN83372.1 CH940651 KRF82319.1 EDW65630.1 KRF82320.1 OUUW01000015 SPP88772.1 CM000162 KRK06433.1 CH933815 EDW08039.1 CH379064 KRT06943.1 EDX01911.1 CH954180 EDV46876.2 EAL32121.2 KQS30224.1 KQS30225.1 CP012528 ALC48584.1 KY407546 ATU47252.1 KRK06435.1 KRG07562.1 KRG07564.1 KRG07565.1 KRG07567.1 CH964239 EDW82571.2 KRT06944.1 KRT06945.1 DS234995 EEB10108.1 KRK06434.1 KRK06436.1 KRG07561.1 AE014298 ACZ95309.1 KRT06942.1 KRG07566.1 M94375 L33831 BT021414 AY070553 KQS30223.1 CH916370 EDW00125.1 CH902644 EDV34801.1 CM000366 EDX18084.1 KPU75290.1 GL888387 EGI61759.1 AAAB01008859 KQ978625 KYN29621.1 KZ288254 PBC30823.1 KQ982080 KYQ60346.1 GFXV01006180 MBW17985.1 KK107064 EZA60894.1 ATLV01012931 ATLV01012932 ATLV01012933 ATLV01012934 ATLV01012935 ATLV01012936 ATLV01012937 ATLV01012938 KE524827 KFB37072.1 ABLF02040171 ABLF02040188 ABLF02040191
Proteomes
UP000005204
UP000053240
UP000218220
UP000283053
UP000007151
UP000053268
+ More
UP000037510 UP000095300 UP000037069 UP000075900 UP000078541 UP000078540 UP000005205 UP000007266 UP000183832 UP000078542 UP000005203 UP000008237 UP000008792 UP000268350 UP000002282 UP000009192 UP000001819 UP000008711 UP000092553 UP000007798 UP000009046 UP000000803 UP000001070 UP000007801 UP000000304 UP000007755 UP000078492 UP000242457 UP000075809 UP000053097 UP000192221 UP000030765 UP000007819
UP000037510 UP000095300 UP000037069 UP000075900 UP000078541 UP000078540 UP000005205 UP000007266 UP000183832 UP000078542 UP000005203 UP000008237 UP000008792 UP000268350 UP000002282 UP000009192 UP000001819 UP000008711 UP000092553 UP000007798 UP000009046 UP000000803 UP000001070 UP000007801 UP000000304 UP000007755 UP000078492 UP000242457 UP000075809 UP000053097 UP000192221 UP000030765 UP000007819
Pfam
Interpro
IPR034182
Kexin/furin
+ More
IPR023828 Peptidase_S8_Ser-AS
IPR022398 Peptidase_S8_His-AS
IPR002884 P_dom
IPR023827 Peptidase_S8_Asp-AS
IPR009030 Growth_fac_rcpt_cys_sf
IPR006212 Furin_repeat
IPR036852 Peptidase_S8/S53_dom_sf
IPR008979 Galactose-bd-like_sf
IPR015500 Peptidase_S8_subtilisin-rel
IPR000209 Peptidase_S8/S53_dom
IPR032778 GF_recep_IV
IPR038466 S8_pro-domain_sf
IPR032815 S8_pro-domain
IPR000742 EGF-like_dom
IPR011031 Multihaem_cyt
IPR023828 Peptidase_S8_Ser-AS
IPR022398 Peptidase_S8_His-AS
IPR002884 P_dom
IPR023827 Peptidase_S8_Asp-AS
IPR009030 Growth_fac_rcpt_cys_sf
IPR006212 Furin_repeat
IPR036852 Peptidase_S8/S53_dom_sf
IPR008979 Galactose-bd-like_sf
IPR015500 Peptidase_S8_subtilisin-rel
IPR000209 Peptidase_S8/S53_dom
IPR032778 GF_recep_IV
IPR038466 S8_pro-domain_sf
IPR032815 S8_pro-domain
IPR000742 EGF-like_dom
IPR011031 Multihaem_cyt
Gene 3D
ProteinModelPortal
H9IYM0
A0A194RGR3
A0A2A4JGI8
A0A2H1WK85
A0A437B8F0
A0A212FLJ6
+ More
A0A194PZ75 A0A0L7LPQ1 A0A2A4K7Y7 A0A212F4A7 Q26489 A0A437BM12 A0A194RFY5 A0A194PYD1 A0A2H1W496 H9IYN1 A0A0A1XGS4 A0A0K8U8C6 A0A0A1X1M3 A0A0L7LUE0 W8APW3 A0A1Q3FTU0 A0A1Q3FU12 A0A1Q3FTV5 A0A1Q3FUD1 T1P9R1 A0A1I8Q1H9 A0A1I8Q1I4 A0A0L0BQD1 A0A2M4DS50 A0A2M3Z0N1 A0A2M4A589 A0A2M4DS69 A0A2M4A5Q5 A0A2M4A4R7 A0A2M4B9F8 A0A2M4B9B2 A0A2M4CJY4 A0A2M4B9D5 Q5DNW1 A0A023EXA8 W4VRF9 A0A336LJ86 A0A336LIK8 A0A2C9GWR4 A0A195FL77 A0A1B6GHW2 A0A195BWS1 A0A1B6CCM9 A0A158NEY8 A0A1Y1KM81 A0A139WNE9 A0A1J1IF38 A0A0C9QJY7 A0A151IEG1 A0A0C9QQN8 A0A088AL19 A0A336LJV0 E2BLN8 A0A0Q9WMJ9 B4M1S6 A0A3B0K7Y0 A0A0R1EG80 B4L8R4 A0A0R3P5B9 B4PXS5 B3NX47 Q29GI8 A0A0Q5T754 A0A0M4ELX6 A0A2D3FB63 A0A0R1EEA9 A0A0Q9XGD4 B4NCL2 A0A0R3P6H0 E0V9N9 A0A0R1EGT9 A0A0Q9XJA6 E1JJL4 A0A0R3P116 A0A0Q9XRF1 P30432 A0A0Q5T532 B4JKM2 B3N0S2 P30432-2 B4R5T4 A0A0P8ZKG0 F4WVD8 A0A1S4GGJ0 A0A195EN83 A0A2A3EGB8 A0A151XIV9 A0A2H8TUQ2 A0A026X0H0 A0A1W4VTY2 A0A1W4VHD4 A0A084VGH8 X1WJ00 A0A1W4VVH0
A0A194PZ75 A0A0L7LPQ1 A0A2A4K7Y7 A0A212F4A7 Q26489 A0A437BM12 A0A194RFY5 A0A194PYD1 A0A2H1W496 H9IYN1 A0A0A1XGS4 A0A0K8U8C6 A0A0A1X1M3 A0A0L7LUE0 W8APW3 A0A1Q3FTU0 A0A1Q3FU12 A0A1Q3FTV5 A0A1Q3FUD1 T1P9R1 A0A1I8Q1H9 A0A1I8Q1I4 A0A0L0BQD1 A0A2M4DS50 A0A2M3Z0N1 A0A2M4A589 A0A2M4DS69 A0A2M4A5Q5 A0A2M4A4R7 A0A2M4B9F8 A0A2M4B9B2 A0A2M4CJY4 A0A2M4B9D5 Q5DNW1 A0A023EXA8 W4VRF9 A0A336LJ86 A0A336LIK8 A0A2C9GWR4 A0A195FL77 A0A1B6GHW2 A0A195BWS1 A0A1B6CCM9 A0A158NEY8 A0A1Y1KM81 A0A139WNE9 A0A1J1IF38 A0A0C9QJY7 A0A151IEG1 A0A0C9QQN8 A0A088AL19 A0A336LJV0 E2BLN8 A0A0Q9WMJ9 B4M1S6 A0A3B0K7Y0 A0A0R1EG80 B4L8R4 A0A0R3P5B9 B4PXS5 B3NX47 Q29GI8 A0A0Q5T754 A0A0M4ELX6 A0A2D3FB63 A0A0R1EEA9 A0A0Q9XGD4 B4NCL2 A0A0R3P6H0 E0V9N9 A0A0R1EGT9 A0A0Q9XJA6 E1JJL4 A0A0R3P116 A0A0Q9XRF1 P30432 A0A0Q5T532 B4JKM2 B3N0S2 P30432-2 B4R5T4 A0A0P8ZKG0 F4WVD8 A0A1S4GGJ0 A0A195EN83 A0A2A3EGB8 A0A151XIV9 A0A2H8TUQ2 A0A026X0H0 A0A1W4VTY2 A0A1W4VHD4 A0A084VGH8 X1WJ00 A0A1W4VVH0
PDB
1P8J
E-value=1.3192e-152,
Score=1388
Ontologies
GO
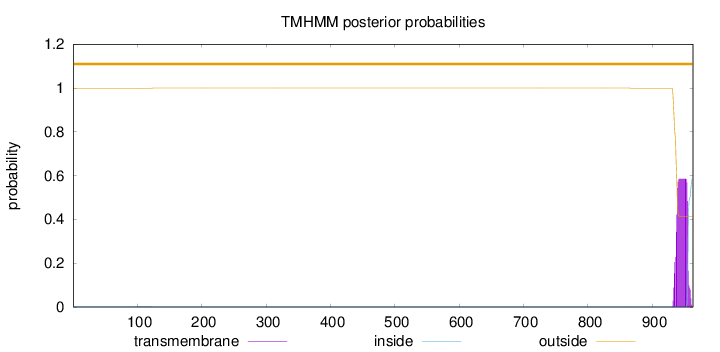
Topology
Subcellular location
Membrane
Length:
963
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.75856
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00118
outside
1 - 963
Population Genetic Test Statistics
Pi
185.508713
Theta
166.357344
Tajima's D
0.06608
CLR
13.924643
CSRT
0.392930353482326
Interpretation
Uncertain
