Gene
KWMTBOMO12425
Pre Gene Modal
BGIBMGA012416
Annotation
nicotinic_acetylcholine_receptor_alpha_8_subunit_[Bombyx_mori]
Full name
Acetylcholine receptor subunit beta-like 2
Alternative Name
Nicotinic acetylcholine receptor beta 2
Location in the cell
PlasmaMembrane Reliability : 4.175
Sequence
CDS
ATGGTGCAGGAGAAAGTAATAATCGTCCGGAGGGTTCCCAACACTTTCTACTATCCGTACAGCTTGTGGCCTATGCCCAAGTTTCAAATCAAAGCTGTTAAACTACTCGAAGCGAATCCTGATGTAAAACGCCTTTACGATGATCTACTTAGCAACTACAATCGGTTGATTCGACCCGTCACTAATGTCAGTGACATATTGACAGTGAGGCTTGGGCTGAAGCTTTCGCAGCTGATGGAGGTTAACCTGAAAAATCAGGTCATGACTACTAACCTTTGGGTGGAGCAGAAATGGTTTGACTATAAATTGCAATGGAACCCAGACGACTACGGAGGAGTGGAAATGCTGTACGTACCGTCGGAGCACATTTGGTTGCCCGACATCGTGCTCTACAACAACTGGGATGGTAATTATGAAGTAACTTTAATGACCAAAGCAACCTTGAAGTACACCGGCGAAGTGAACTGGAAGCCGCCAGCGATCTACAAATCCTCCTGCGAGATCAACGTCGAATACTTTCCATTTGATGAACAAACATGCTTCATGAAGTTTGGTTCTTGGACCTATAATGGAGCTCAGGTTGACCTTAAACATATGGACCAGTCTTCCGGAAGCAGTCTAGTTCACGTCGGTATTGATCTAAGCGAGTTTTATTTGTCAGTCGAGTGGGATATACTTGAAGTACCGGCTACGAGGAACGAAGAGTACTACCCTTGTTGCCCCGAGCCTTTTTCTGACATAACATTCAAACTGACGATGCGCAGAAAAACATTGTTCTATACCGTCAACCTGATTATACCCTGTGTGGGTTTGACGTTTTTGACCGTCTTAGTTTTCTATCTACCCTCGGATTCGGGTGAGAAGATTTCGCTCTGCATCTCCATCCTGGTGTCCCTCACTGTGTTCTTCCTCGGTTTGGCCGAGATCATCCCGCCAACATCCCTGGCAATCCCATTGCTCGGGAAATATCTTCTATTCACCATGATACTCGTCTCGCTCAGCGTATTGGTCACTGTGTGCATAGTAAACGTGCATTTCAGGTCTCCGTCAACGCACACAATGTCGCCGTGGATGAAGAAGTTATTCCTTCAGCTGATGCCCAAACTCTTGATGATGAGGAGGACGAAGTACTCGCTTCCAGACTACGATGACACGTTCGTATCTAATGGATACACCAACGAATTGGAAATGAGTCGAGATAGTTTGACTGACGCTTTCGGAGATTCGAAATGTGACAACGGAGATTACCGCAAGTCTCCTGCTCCTGAAGACGACATGATGGGAGCCAGCGCACACCAGAGACCTTCAGTAACCGAATCTGAGAATATGCTGCCTCGCCATTTGTCGCCTGAAGTCGCTGCTGCCCTGAAGAGTGTCAGATTCATCGCTCAACACATTAAAGATGCTGACAAGGATAATGAGGTCATCGAAGATTGGAAGTTCATGTCAATGGTGCTGGACCGCTTCTTTCTCTGGCTGTTCACCATAGCTTGTTTTGTGGGAACTTTTGGGATAATCTTCCAATCCCCGTCTCTATACGACACCAGAGTCCCTGTCGATCAGCAAATATCATCTATACCGATGAAGAAAAACAACTTTTTCTATCCTAAAGACATTGAAACTATTGGAATTATTAGTTGA
Protein
MVQEKVIIVRRVPNTFYYPYSLWPMPKFQIKAVKLLEANPDVKRLYDDLLSNYNRLIRPVTNVSDILTVRLGLKLSQLMEVNLKNQVMTTNLWVEQKWFDYKLQWNPDDYGGVEMLYVPSEHIWLPDIVLYNNWDGNYEVTLMTKATLKYTGEVNWKPPAIYKSSCEINVEYFPFDEQTCFMKFGSWTYNGAQVDLKHMDQSSGSSLVHVGIDLSEFYLSVEWDILEVPATRNEEYYPCCPEPFSDITFKLTMRRKTLFYTVNLIIPCVGLTFLTVLVFYLPSDSGEKISLCISILVSLTVFFLGLAEIIPPTSLAIPLLGKYLLFTMILVSLSVLVTVCIVNVHFRSPSTHTMSPWMKKLFLQLMPKLLMMRRTKYSLPDYDDTFVSNGYTNELEMSRDSLTDAFGDSKCDNGDYRKSPAPEDDMMGASAHQRPSVTESENMLPRHLSPEVAAALKSVRFIAQHIKDADKDNEVIEDWKFMSMVLDRFFLWLFTIACFVGTFGIIFQSPSLYDTRVPVDQQISSIPMKKNNFFYPKDIETIGIIS
Summary
Description
After binding acetylcholine, the AChR responds by an extensive change in conformation that affects all subunits and leads to opening of an ion-conducting channel across the plasma membrane.
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family. Acetylcholine receptor (TC 1.A.9.1) subfamily.
Belongs to the ligand-gated ion channel (TC 1.A.9) family. Acetylcholine receptor (TC 1.A.9.1) subfamily.
Keywords
Alternative splicing
Cell junction
Cell membrane
Complete proteome
Disulfide bond
Glycoprotein
Ion channel
Ion transport
Ligand-gated ion channel
Membrane
Postsynaptic cell membrane
Receptor
Reference proteome
RNA editing
Signal
Synapse
Transmembrane
Transmembrane helix
Transport
Feature
chain Acetylcholine receptor subunit beta-like 2
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A8HTD0
A8CGN2
A8CGM9
B5LVL4
A0A2A4K147
O96632
+ More
A0A0H4TM43 Q8WRS1 A0A0H4TME7 A0A0A7RWN6 A0A194PWH7 H9JSA5 A0A1B0DGT5 A0A182XWD4 A0A1Y9HEI1 Q7PQY4 A0A1W7R7Y4 A0A1S4G816 A0A1B0D5D3 A0A0R3NNI9 A0A1Q3FVK1 Q297E0 B4GE89 A0A3B0K5L7 B4NF13 A0A1W4V4C7 B4QT71 B4PVT5 B3N1R1 B3P727 P25162 W8C655 B4HH28 A0A0A1WKB2 A0A0K8UFQ9 B4JFZ8 A0A0M4ENR1 B4K9Y1 A0A034WMD6 A0A0Q9X0L2 A0A1B6CSI7 B4LXX6 A0A1B6LB43 H6TY12 A0A0K1U4G4 W6E9B6 A0A1A9X9A0 W6EAG0 B7UDC9 K7IUV1 A0A1I8P470 O46133 A0A348BSW5 A0A232F686 A0A1I8MQQ8 A0A2Z4N5Q9 Q8MUR0 D3UA20 D2CVP0 A0A0L0BP39 T1PH14 F4WAV6 A0A1I8P432 A0A2A3EKN1 A0A158P3L7 W6EL46 W6E971 A0A0N0U641 A8DIV5 D6WB25 C3TS66 A0A075W270 A0A195F170 C3TS54 A0A139WNS9 A0A154P3T9 A0A0L7R4A8 A0A139WNI5 A8DIU4 C3TS55 B5ARH0 A0A195CSV5 A0A026WYS6 A0A1Y1LP14 T2HNV7 A0A1W4XG06 E0VDF4 Q5F2K5 Q1NZ58 Q4ZHN1 Q4ZHN0 A0A1D2MEM2 A0A067RUF6 A0A2H1WTV6 A0A151JA70 A0A2S2R818 A0A182P6N9 A0A182HU27 A0A182VYJ2 A0A182NDW0
A0A0H4TM43 Q8WRS1 A0A0H4TME7 A0A0A7RWN6 A0A194PWH7 H9JSA5 A0A1B0DGT5 A0A182XWD4 A0A1Y9HEI1 Q7PQY4 A0A1W7R7Y4 A0A1S4G816 A0A1B0D5D3 A0A0R3NNI9 A0A1Q3FVK1 Q297E0 B4GE89 A0A3B0K5L7 B4NF13 A0A1W4V4C7 B4QT71 B4PVT5 B3N1R1 B3P727 P25162 W8C655 B4HH28 A0A0A1WKB2 A0A0K8UFQ9 B4JFZ8 A0A0M4ENR1 B4K9Y1 A0A034WMD6 A0A0Q9X0L2 A0A1B6CSI7 B4LXX6 A0A1B6LB43 H6TY12 A0A0K1U4G4 W6E9B6 A0A1A9X9A0 W6EAG0 B7UDC9 K7IUV1 A0A1I8P470 O46133 A0A348BSW5 A0A232F686 A0A1I8MQQ8 A0A2Z4N5Q9 Q8MUR0 D3UA20 D2CVP0 A0A0L0BP39 T1PH14 F4WAV6 A0A1I8P432 A0A2A3EKN1 A0A158P3L7 W6EL46 W6E971 A0A0N0U641 A8DIV5 D6WB25 C3TS66 A0A075W270 A0A195F170 C3TS54 A0A139WNS9 A0A154P3T9 A0A0L7R4A8 A0A139WNI5 A8DIU4 C3TS55 B5ARH0 A0A195CSV5 A0A026WYS6 A0A1Y1LP14 T2HNV7 A0A1W4XG06 E0VDF4 Q5F2K5 Q1NZ58 Q4ZHN1 Q4ZHN0 A0A1D2MEM2 A0A067RUF6 A0A2H1WTV6 A0A151JA70 A0A2S2R818 A0A182P6N9 A0A182HU27 A0A182VYJ2 A0A182NDW0
Pubmed
17868469
25504620
26354079
19121390
25244985
12364791
+ More
14747013 15676276 17210077 15632085 17994087 17550304 2121539 9109505 10731132 12537572 12537569 12907802 24495485 25830018 25348373 25701599 20075255 9660807 28648823 25315136 29136191 12752659 20087392 26108605 21719571 21347285 17880682 18362917 19820115 19320762 19533628 24508170 28004739 20566863 27289101 24845553
14747013 15676276 17210077 15632085 17994087 17550304 2121539 9109505 10731132 12537572 12537569 12907802 24495485 25830018 25348373 25701599 20075255 9660807 28648823 25315136 29136191 12752659 20087392 26108605 21719571 21347285 17880682 18362917 19820115 19320762 19533628 24508170 28004739 20566863 27289101 24845553
EMBL
EU143799
ABV72690.1
EU082086
ABV45522.1
EU082085
ABV45521.1
+ More
EU914853 ACG76113.1 NWSH01000256 PCG77967.1 AF096879 AAD09809.1 KP711057 RSAL01000243 AKQ12753.1 RVE43612.1 AF418987 AAL40742.1 KP711050 AKQ12746.1 KP101247 AJA39823.1 KQ459591 KPI97114.1 BABH01033800 BABH01033801 BABH01033802 BABH01033803 AJVK01060318 AY705403 AAAB01008859 AAU12512.1 EAA08126.4 GEHC01000367 JAV47278.1 AJVK01025143 CM000070 KRT00611.1 GFDL01003573 JAV31472.1 EAL28266.3 CH479182 EDW33924.1 OUUW01000007 SPP83340.1 CH964251 EDW83388.1 CM000364 EDX14235.1 CM000160 EDW98934.1 CH902661 EDV34030.2 CH954182 EDV53847.1 X55676 Y14678 AE014297 AY089498 GAMC01008706 JAB97849.1 CH480815 EDW43504.1 GBXI01015322 JAC98969.1 GDHF01026898 GDHF01003421 JAI25416.1 JAI48893.1 CH916369 EDV92537.1 CP012526 ALC47302.1 CH933806 EDW15629.1 GAKP01003143 JAC55809.1 KRG01648.1 GEDC01020789 JAS16509.1 CH940650 EDW66842.1 GEBQ01019064 JAT20913.1 JN390947 AFA28130.1 KP725466 AKV94623.1 KF873590 AHJ11219.1 KF873589 AHJ11218.1 FJ481979 ACK75719.1 AAZX01002653 AJ000391 CAA04053.1 LC389189 BBE49568.1 NNAY01000907 OXU25983.1 MF612139 QKKF02026599 AWX65627.1 RZF36327.1 AF514804 KJ939595 AAM51823.1 AJE70266.1 FJ821439 ACY82693.1 EU879964 ACJ64922.1 JRES01001582 KNC21743.1 KA648102 AFP62731.1 GL888053 EGI68712.1 KZ288229 PBC31729.1 ADTU01008200 KF873592 AHJ11221.1 KF873591 AHJ11220.1 KQ435756 KOX75921.1 EF526093 ABS86915.1 KQ971311 EEZ99341.2 EU937809 ACP31314.1 KJ747359 AIG92772.1 KQ981880 KYN33917.1 EU937797 EU937799 EU937800 EU937801 ACP31302.1 KYB29425.1 KQ434809 KZC06609.1 KQ414658 KOC65659.1 KYB29426.1 EF526090 ABS86912.1 EU937798 ACP31303.1 EU871526 ACG49259.1 KQ977304 KYN03783.1 KK107064 EZA60916.1 GEZM01051844 JAV74671.1 AB748924 BAN81842.1 DS235075 EEB11410.1 AJ880082 CAI54099.1 DQ480159 ABE67099.1 DQ009491 AAY28924.1 DQ009492 AAY28925.1 LJIJ01001600 ODM91301.1 KK852460 KDR23474.1 ODYU01010968 SOQ56406.1 KQ979321 KYN21946.1 GGMS01016915 MBY86118.1 APCN01000602 APCN01000603
EU914853 ACG76113.1 NWSH01000256 PCG77967.1 AF096879 AAD09809.1 KP711057 RSAL01000243 AKQ12753.1 RVE43612.1 AF418987 AAL40742.1 KP711050 AKQ12746.1 KP101247 AJA39823.1 KQ459591 KPI97114.1 BABH01033800 BABH01033801 BABH01033802 BABH01033803 AJVK01060318 AY705403 AAAB01008859 AAU12512.1 EAA08126.4 GEHC01000367 JAV47278.1 AJVK01025143 CM000070 KRT00611.1 GFDL01003573 JAV31472.1 EAL28266.3 CH479182 EDW33924.1 OUUW01000007 SPP83340.1 CH964251 EDW83388.1 CM000364 EDX14235.1 CM000160 EDW98934.1 CH902661 EDV34030.2 CH954182 EDV53847.1 X55676 Y14678 AE014297 AY089498 GAMC01008706 JAB97849.1 CH480815 EDW43504.1 GBXI01015322 JAC98969.1 GDHF01026898 GDHF01003421 JAI25416.1 JAI48893.1 CH916369 EDV92537.1 CP012526 ALC47302.1 CH933806 EDW15629.1 GAKP01003143 JAC55809.1 KRG01648.1 GEDC01020789 JAS16509.1 CH940650 EDW66842.1 GEBQ01019064 JAT20913.1 JN390947 AFA28130.1 KP725466 AKV94623.1 KF873590 AHJ11219.1 KF873589 AHJ11218.1 FJ481979 ACK75719.1 AAZX01002653 AJ000391 CAA04053.1 LC389189 BBE49568.1 NNAY01000907 OXU25983.1 MF612139 QKKF02026599 AWX65627.1 RZF36327.1 AF514804 KJ939595 AAM51823.1 AJE70266.1 FJ821439 ACY82693.1 EU879964 ACJ64922.1 JRES01001582 KNC21743.1 KA648102 AFP62731.1 GL888053 EGI68712.1 KZ288229 PBC31729.1 ADTU01008200 KF873592 AHJ11221.1 KF873591 AHJ11220.1 KQ435756 KOX75921.1 EF526093 ABS86915.1 KQ971311 EEZ99341.2 EU937809 ACP31314.1 KJ747359 AIG92772.1 KQ981880 KYN33917.1 EU937797 EU937799 EU937800 EU937801 ACP31302.1 KYB29425.1 KQ434809 KZC06609.1 KQ414658 KOC65659.1 KYB29426.1 EF526090 ABS86912.1 EU937798 ACP31303.1 EU871526 ACG49259.1 KQ977304 KYN03783.1 KK107064 EZA60916.1 GEZM01051844 JAV74671.1 AB748924 BAN81842.1 DS235075 EEB11410.1 AJ880082 CAI54099.1 DQ480159 ABE67099.1 DQ009491 AAY28924.1 DQ009492 AAY28925.1 LJIJ01001600 ODM91301.1 KK852460 KDR23474.1 ODYU01010968 SOQ56406.1 KQ979321 KYN21946.1 GGMS01016915 MBY86118.1 APCN01000602 APCN01000603
Proteomes
UP000218220
UP000283053
UP000053268
UP000005204
UP000092462
UP000076408
+ More
UP000075900 UP000007062 UP000001819 UP000008744 UP000268350 UP000007798 UP000192221 UP000000304 UP000002282 UP000007801 UP000008711 UP000000803 UP000001292 UP000001070 UP000092553 UP000009192 UP000008792 UP000092443 UP000002358 UP000095300 UP000215335 UP000095301 UP000291343 UP000037069 UP000007755 UP000242457 UP000005205 UP000053105 UP000007266 UP000078541 UP000076502 UP000053825 UP000078542 UP000053097 UP000192223 UP000009046 UP000094527 UP000027135 UP000078492 UP000075885 UP000075840 UP000075920 UP000075884
UP000075900 UP000007062 UP000001819 UP000008744 UP000268350 UP000007798 UP000192221 UP000000304 UP000002282 UP000007801 UP000008711 UP000000803 UP000001292 UP000001070 UP000092553 UP000009192 UP000008792 UP000092443 UP000002358 UP000095300 UP000215335 UP000095301 UP000291343 UP000037069 UP000007755 UP000242457 UP000005205 UP000053105 UP000007266 UP000078541 UP000076502 UP000053825 UP000078542 UP000053097 UP000192223 UP000009046 UP000094527 UP000027135 UP000078492 UP000075885 UP000075840 UP000075920 UP000075884
Interpro
Gene 3D
ProteinModelPortal
A8HTD0
A8CGN2
A8CGM9
B5LVL4
A0A2A4K147
O96632
+ More
A0A0H4TM43 Q8WRS1 A0A0H4TME7 A0A0A7RWN6 A0A194PWH7 H9JSA5 A0A1B0DGT5 A0A182XWD4 A0A1Y9HEI1 Q7PQY4 A0A1W7R7Y4 A0A1S4G816 A0A1B0D5D3 A0A0R3NNI9 A0A1Q3FVK1 Q297E0 B4GE89 A0A3B0K5L7 B4NF13 A0A1W4V4C7 B4QT71 B4PVT5 B3N1R1 B3P727 P25162 W8C655 B4HH28 A0A0A1WKB2 A0A0K8UFQ9 B4JFZ8 A0A0M4ENR1 B4K9Y1 A0A034WMD6 A0A0Q9X0L2 A0A1B6CSI7 B4LXX6 A0A1B6LB43 H6TY12 A0A0K1U4G4 W6E9B6 A0A1A9X9A0 W6EAG0 B7UDC9 K7IUV1 A0A1I8P470 O46133 A0A348BSW5 A0A232F686 A0A1I8MQQ8 A0A2Z4N5Q9 Q8MUR0 D3UA20 D2CVP0 A0A0L0BP39 T1PH14 F4WAV6 A0A1I8P432 A0A2A3EKN1 A0A158P3L7 W6EL46 W6E971 A0A0N0U641 A8DIV5 D6WB25 C3TS66 A0A075W270 A0A195F170 C3TS54 A0A139WNS9 A0A154P3T9 A0A0L7R4A8 A0A139WNI5 A8DIU4 C3TS55 B5ARH0 A0A195CSV5 A0A026WYS6 A0A1Y1LP14 T2HNV7 A0A1W4XG06 E0VDF4 Q5F2K5 Q1NZ58 Q4ZHN1 Q4ZHN0 A0A1D2MEM2 A0A067RUF6 A0A2H1WTV6 A0A151JA70 A0A2S2R818 A0A182P6N9 A0A182HU27 A0A182VYJ2 A0A182NDW0
A0A0H4TM43 Q8WRS1 A0A0H4TME7 A0A0A7RWN6 A0A194PWH7 H9JSA5 A0A1B0DGT5 A0A182XWD4 A0A1Y9HEI1 Q7PQY4 A0A1W7R7Y4 A0A1S4G816 A0A1B0D5D3 A0A0R3NNI9 A0A1Q3FVK1 Q297E0 B4GE89 A0A3B0K5L7 B4NF13 A0A1W4V4C7 B4QT71 B4PVT5 B3N1R1 B3P727 P25162 W8C655 B4HH28 A0A0A1WKB2 A0A0K8UFQ9 B4JFZ8 A0A0M4ENR1 B4K9Y1 A0A034WMD6 A0A0Q9X0L2 A0A1B6CSI7 B4LXX6 A0A1B6LB43 H6TY12 A0A0K1U4G4 W6E9B6 A0A1A9X9A0 W6EAG0 B7UDC9 K7IUV1 A0A1I8P470 O46133 A0A348BSW5 A0A232F686 A0A1I8MQQ8 A0A2Z4N5Q9 Q8MUR0 D3UA20 D2CVP0 A0A0L0BP39 T1PH14 F4WAV6 A0A1I8P432 A0A2A3EKN1 A0A158P3L7 W6EL46 W6E971 A0A0N0U641 A8DIV5 D6WB25 C3TS66 A0A075W270 A0A195F170 C3TS54 A0A139WNS9 A0A154P3T9 A0A0L7R4A8 A0A139WNI5 A8DIU4 C3TS55 B5ARH0 A0A195CSV5 A0A026WYS6 A0A1Y1LP14 T2HNV7 A0A1W4XG06 E0VDF4 Q5F2K5 Q1NZ58 Q4ZHN1 Q4ZHN0 A0A1D2MEM2 A0A067RUF6 A0A2H1WTV6 A0A151JA70 A0A2S2R818 A0A182P6N9 A0A182HU27 A0A182VYJ2 A0A182NDW0
PDB
6CNK
E-value=6.12191e-104,
Score=966
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell junction
Synapse
Postsynaptic cell membrane
Cell membrane
Synapse
Postsynaptic cell membrane
Cell membrane
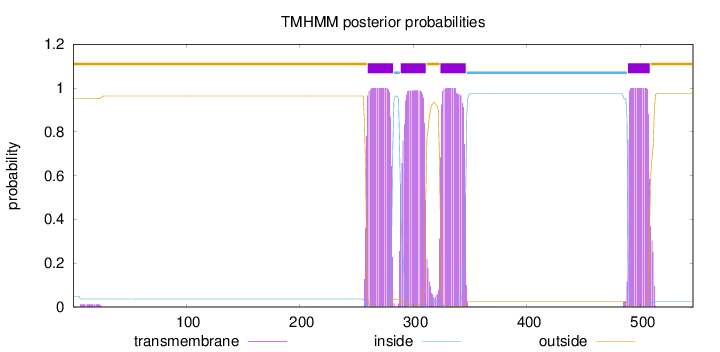
Length:
546
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
87.71248
Exp number, first 60 AAs:
0.23342
Total prob of N-in:
0.04852
outside
1 - 259
TMhelix
260 - 282
inside
283 - 288
TMhelix
289 - 311
outside
312 - 323
TMhelix
324 - 346
inside
347 - 488
TMhelix
489 - 508
outside
509 - 546
Population Genetic Test Statistics
Pi
195.118359
Theta
177.934567
Tajima's D
0.297407
CLR
0.468408
CSRT
0.447927603619819
Interpretation
Uncertain
