Gene
KWMTBOMO12422
Annotation
PREDICTED:_uncharacterized_protein_LOC105841556_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.826 PlasmaMembrane Reliability : 1.627
Sequence
CDS
ATGGTTTTCGTTACCCTGTCAATACCCCTTCACCCGGAAACAACGGTCAGCGTCGCATGGTTCTTCGAAGCTAATTACTACAATGTTGACAATTCCACCTATTTCGAACCGCTCTTAGGAGATATAAATATCGCAAGGAGCCGTAACGAACGTAATGCCGATTTGATGAGTGATTTCACGAGGCGCAGACTTTATTTCCTCATTGAAAGAGTACTGGAACAACACGACTACAGTGGACGGGCGTGCTTATTGAGGGTCATTTGTGAGAACGGAACTTCCAATCTGTTGCATAGCGGTGTTTTAGGAGATATACTGCATTTGATACTAACTCCTTCCACATCTATGGCCGAAGACAATCTACCCGACTGTTACTATGAAGCCGAATATTATGGTTTGGATGATCGTTGCGAAAAATACACGGAATCGTGCCCATCTAGTCCTTTAGAAGTGATCACGACATTTATTGACCGATTAATATAA
Protein
MVFVTLSIPLHPETTVSVAWFFEANYYNVDNSTYFEPLLGDINIARSRNERNADLMSDFTRRRLYFLIERVLEQHDYSGRACLLRVICENGTSNLLHSGVLGDILHLILTPSTSMAEDNLPDCYYEAEYYGLDDRCEKYTESCPSSPLEVITTFIDRLI
Summary
Uniprot
A0A2A4IYV4
A0A3S2LIT6
A0A2H1WT81
A0A194RAX1
A0A158NBC1
A0A3L8DNQ1
+ More
A0A194PVU1 A0A3S2LTK5 A0A212FII2 A0A232EV53 A0A151WZT8 A0A182Q2Q2 K7JHN0 A0A195DZ00 A0A151IPR7 A0A2H1V773 F4WCU8 E1ZY84 E1ZY87 A0A195EVR7 Q16Y12 A0A182KMN1 A0A1I8JWC0 Q7Q267 A0A154P2C3 E9IZZ0 A0A182VJ92 A0A182JF75 A0A182UKJ9 A0A182UIE5 A0A182LRB5 A0A1W4X9C3 A0A1I8JW78 A7UV00 A0A026X0N3 Q16Y13 A0A182S8E1 A0A182KMN0 A0A151IPT0 A0A182VXL8 W5J5Z8 A0A182QYQ4 A0A1B0BFM1 A0A2A3EQY2 A0A1A9ZWZ2 A0A1A9VQA2 A0A1S4FK67 A0A232EV44 A0A182G5Y7 A0A2A4K1R2 A0A0Q9X7U9 A0A1A9YLR9 A0A1J1HU84 A0A1B0G4A5 A0A182YID5 F4WCU7 A0A195BRK3 A0A182P8Y0 A0A151X0C6 A0A195EVP8 A0A0Q9W574 A0A182RBF1 A0A195DZB5 A0A182F823 A0A182N6E5 A0A182GGB9 A0A084VWK2 A0A0Q9X810 A0A0C9RFI2 D6W7Y6 Q16PN3 A0NFN4 A0A182IJ47 A0A0L7QSD8 A0A182KMP1 D6W795 A0A1B0CF03 A0A087ZVG9 A0A182H5B0 A0A1B6I4G2 A0A1Y9J095 B0WP61 A0A182F824 A0A0A1XR24 A0A1I8NWF6 B4LQ95 H9JSD6 A0A3L8DPG8 A0A154P435 E9IZZ1 A0A026X0L1 A0A0T6B3Y2 A0A182YID0 A0A336MZT0
A0A194PVU1 A0A3S2LTK5 A0A212FII2 A0A232EV53 A0A151WZT8 A0A182Q2Q2 K7JHN0 A0A195DZ00 A0A151IPR7 A0A2H1V773 F4WCU8 E1ZY84 E1ZY87 A0A195EVR7 Q16Y12 A0A182KMN1 A0A1I8JWC0 Q7Q267 A0A154P2C3 E9IZZ0 A0A182VJ92 A0A182JF75 A0A182UKJ9 A0A182UIE5 A0A182LRB5 A0A1W4X9C3 A0A1I8JW78 A7UV00 A0A026X0N3 Q16Y13 A0A182S8E1 A0A182KMN0 A0A151IPT0 A0A182VXL8 W5J5Z8 A0A182QYQ4 A0A1B0BFM1 A0A2A3EQY2 A0A1A9ZWZ2 A0A1A9VQA2 A0A1S4FK67 A0A232EV44 A0A182G5Y7 A0A2A4K1R2 A0A0Q9X7U9 A0A1A9YLR9 A0A1J1HU84 A0A1B0G4A5 A0A182YID5 F4WCU7 A0A195BRK3 A0A182P8Y0 A0A151X0C6 A0A195EVP8 A0A0Q9W574 A0A182RBF1 A0A195DZB5 A0A182F823 A0A182N6E5 A0A182GGB9 A0A084VWK2 A0A0Q9X810 A0A0C9RFI2 D6W7Y6 Q16PN3 A0NFN4 A0A182IJ47 A0A0L7QSD8 A0A182KMP1 D6W795 A0A1B0CF03 A0A087ZVG9 A0A182H5B0 A0A1B6I4G2 A0A1Y9J095 B0WP61 A0A182F824 A0A0A1XR24 A0A1I8NWF6 B4LQ95 H9JSD6 A0A3L8DPG8 A0A154P435 E9IZZ1 A0A026X0L1 A0A0T6B3Y2 A0A182YID0 A0A336MZT0
Pubmed
EMBL
NWSH01005203
PCG64360.1
RSAL01000098
RVE47597.1
ODYU01010878
SOQ56258.1
+ More
KQ460473 KPJ14430.1 ADTU01010843 ADTU01010844 QOIP01000006 RLU21509.1 KQ459591 KPI97113.1 RSAL01000243 RVE43611.1 AGBW02008386 OWR53528.1 NNAY01002055 OXU22223.1 KQ982630 KYQ53415.1 AXCN02000285 KQ980050 KYN18056.1 KQ976812 KYN08162.1 ODYU01001042 SOQ36700.1 GL888074 EGI67929.1 GL435204 EFN73783.1 EFN73786.1 KQ981953 KYN32333.1 CH477528 EAT39491.1 AAAB01008978 EAA13624.5 KQ434804 KZC06085.1 GL767291 EFZ13820.1 AXCP01007617 AXCM01004116 EDO63475.2 KK107039 EZA61847.1 EAT39490.1 KYN08161.1 ADMH02002112 ETN58823.1 JXJN01013570 KZ288193 PBC34175.1 OXU22222.1 JXUM01146977 KQ570273 KXJ68403.1 NWSH01000256 PCG77966.1 CH933808 KRG04427.1 CVRI01000021 CRK91563.1 CCAG010004889 EGI67928.1 KQ976417 KYM89667.1 KYQ53416.1 KYN32335.1 CH940648 KRF79970.1 KYN18057.1 JXUM01001538 JXUM01001539 JXUM01001540 JXUM01001541 JXUM01001542 JXUM01001543 JXUM01001544 KQ560122 KXJ84362.1 ATLV01017623 KE525179 KFB42346.1 KRG04428.1 GBYB01011882 JAG81649.1 KQ971307 EFA11156.1 CH477776 EAT36323.1 EAU76181.1 KQ414768 KOC61376.1 EFA11041.2 AJWK01009488 JXUM01026119 KQ560745 KXJ81033.1 GECU01025875 JAS81831.1 DS232020 EDS32106.1 GBXI01001259 JAD13033.1 EDW61380.2 BABH01033796 BABH01033797 RLU22186.1 KZC06094.1 EFZ13874.1 EZA61845.1 LJIG01009941 KRT82073.1 UFQS01003180 UFQT01002900 UFQT01003180 SSX15325.1 SSX34241.1 SSX34699.1
KQ460473 KPJ14430.1 ADTU01010843 ADTU01010844 QOIP01000006 RLU21509.1 KQ459591 KPI97113.1 RSAL01000243 RVE43611.1 AGBW02008386 OWR53528.1 NNAY01002055 OXU22223.1 KQ982630 KYQ53415.1 AXCN02000285 KQ980050 KYN18056.1 KQ976812 KYN08162.1 ODYU01001042 SOQ36700.1 GL888074 EGI67929.1 GL435204 EFN73783.1 EFN73786.1 KQ981953 KYN32333.1 CH477528 EAT39491.1 AAAB01008978 EAA13624.5 KQ434804 KZC06085.1 GL767291 EFZ13820.1 AXCP01007617 AXCM01004116 EDO63475.2 KK107039 EZA61847.1 EAT39490.1 KYN08161.1 ADMH02002112 ETN58823.1 JXJN01013570 KZ288193 PBC34175.1 OXU22222.1 JXUM01146977 KQ570273 KXJ68403.1 NWSH01000256 PCG77966.1 CH933808 KRG04427.1 CVRI01000021 CRK91563.1 CCAG010004889 EGI67928.1 KQ976417 KYM89667.1 KYQ53416.1 KYN32335.1 CH940648 KRF79970.1 KYN18057.1 JXUM01001538 JXUM01001539 JXUM01001540 JXUM01001541 JXUM01001542 JXUM01001543 JXUM01001544 KQ560122 KXJ84362.1 ATLV01017623 KE525179 KFB42346.1 KRG04428.1 GBYB01011882 JAG81649.1 KQ971307 EFA11156.1 CH477776 EAT36323.1 EAU76181.1 KQ414768 KOC61376.1 EFA11041.2 AJWK01009488 JXUM01026119 KQ560745 KXJ81033.1 GECU01025875 JAS81831.1 DS232020 EDS32106.1 GBXI01001259 JAD13033.1 EDW61380.2 BABH01033796 BABH01033797 RLU22186.1 KZC06094.1 EFZ13874.1 EZA61845.1 LJIG01009941 KRT82073.1 UFQS01003180 UFQT01002900 UFQT01003180 SSX15325.1 SSX34241.1 SSX34699.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000005205
UP000279307
UP000053268
+ More
UP000007151 UP000215335 UP000075809 UP000075886 UP000002358 UP000078492 UP000078542 UP000007755 UP000000311 UP000078541 UP000008820 UP000075882 UP000076407 UP000007062 UP000076502 UP000075903 UP000075880 UP000075902 UP000075883 UP000192223 UP000053097 UP000075901 UP000075920 UP000000673 UP000092460 UP000242457 UP000092445 UP000078200 UP000069940 UP000249989 UP000009192 UP000092443 UP000183832 UP000092444 UP000076408 UP000078540 UP000075885 UP000008792 UP000075900 UP000069272 UP000075884 UP000030765 UP000007266 UP000053825 UP000092461 UP000005203 UP000002320 UP000095300 UP000005204
UP000007151 UP000215335 UP000075809 UP000075886 UP000002358 UP000078492 UP000078542 UP000007755 UP000000311 UP000078541 UP000008820 UP000075882 UP000076407 UP000007062 UP000076502 UP000075903 UP000075880 UP000075902 UP000075883 UP000192223 UP000053097 UP000075901 UP000075920 UP000000673 UP000092460 UP000242457 UP000092445 UP000078200 UP000069940 UP000249989 UP000009192 UP000092443 UP000183832 UP000092444 UP000076408 UP000078540 UP000075885 UP000008792 UP000075900 UP000069272 UP000075884 UP000030765 UP000007266 UP000053825 UP000092461 UP000005203 UP000002320 UP000095300 UP000005204
PRIDE
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
A0A2A4IYV4
A0A3S2LIT6
A0A2H1WT81
A0A194RAX1
A0A158NBC1
A0A3L8DNQ1
+ More
A0A194PVU1 A0A3S2LTK5 A0A212FII2 A0A232EV53 A0A151WZT8 A0A182Q2Q2 K7JHN0 A0A195DZ00 A0A151IPR7 A0A2H1V773 F4WCU8 E1ZY84 E1ZY87 A0A195EVR7 Q16Y12 A0A182KMN1 A0A1I8JWC0 Q7Q267 A0A154P2C3 E9IZZ0 A0A182VJ92 A0A182JF75 A0A182UKJ9 A0A182UIE5 A0A182LRB5 A0A1W4X9C3 A0A1I8JW78 A7UV00 A0A026X0N3 Q16Y13 A0A182S8E1 A0A182KMN0 A0A151IPT0 A0A182VXL8 W5J5Z8 A0A182QYQ4 A0A1B0BFM1 A0A2A3EQY2 A0A1A9ZWZ2 A0A1A9VQA2 A0A1S4FK67 A0A232EV44 A0A182G5Y7 A0A2A4K1R2 A0A0Q9X7U9 A0A1A9YLR9 A0A1J1HU84 A0A1B0G4A5 A0A182YID5 F4WCU7 A0A195BRK3 A0A182P8Y0 A0A151X0C6 A0A195EVP8 A0A0Q9W574 A0A182RBF1 A0A195DZB5 A0A182F823 A0A182N6E5 A0A182GGB9 A0A084VWK2 A0A0Q9X810 A0A0C9RFI2 D6W7Y6 Q16PN3 A0NFN4 A0A182IJ47 A0A0L7QSD8 A0A182KMP1 D6W795 A0A1B0CF03 A0A087ZVG9 A0A182H5B0 A0A1B6I4G2 A0A1Y9J095 B0WP61 A0A182F824 A0A0A1XR24 A0A1I8NWF6 B4LQ95 H9JSD6 A0A3L8DPG8 A0A154P435 E9IZZ1 A0A026X0L1 A0A0T6B3Y2 A0A182YID0 A0A336MZT0
A0A194PVU1 A0A3S2LTK5 A0A212FII2 A0A232EV53 A0A151WZT8 A0A182Q2Q2 K7JHN0 A0A195DZ00 A0A151IPR7 A0A2H1V773 F4WCU8 E1ZY84 E1ZY87 A0A195EVR7 Q16Y12 A0A182KMN1 A0A1I8JWC0 Q7Q267 A0A154P2C3 E9IZZ0 A0A182VJ92 A0A182JF75 A0A182UKJ9 A0A182UIE5 A0A182LRB5 A0A1W4X9C3 A0A1I8JW78 A7UV00 A0A026X0N3 Q16Y13 A0A182S8E1 A0A182KMN0 A0A151IPT0 A0A182VXL8 W5J5Z8 A0A182QYQ4 A0A1B0BFM1 A0A2A3EQY2 A0A1A9ZWZ2 A0A1A9VQA2 A0A1S4FK67 A0A232EV44 A0A182G5Y7 A0A2A4K1R2 A0A0Q9X7U9 A0A1A9YLR9 A0A1J1HU84 A0A1B0G4A5 A0A182YID5 F4WCU7 A0A195BRK3 A0A182P8Y0 A0A151X0C6 A0A195EVP8 A0A0Q9W574 A0A182RBF1 A0A195DZB5 A0A182F823 A0A182N6E5 A0A182GGB9 A0A084VWK2 A0A0Q9X810 A0A0C9RFI2 D6W7Y6 Q16PN3 A0NFN4 A0A182IJ47 A0A0L7QSD8 A0A182KMP1 D6W795 A0A1B0CF03 A0A087ZVG9 A0A182H5B0 A0A1B6I4G2 A0A1Y9J095 B0WP61 A0A182F824 A0A0A1XR24 A0A1I8NWF6 B4LQ95 H9JSD6 A0A3L8DPG8 A0A154P435 E9IZZ1 A0A026X0L1 A0A0T6B3Y2 A0A182YID0 A0A336MZT0
Ontologies
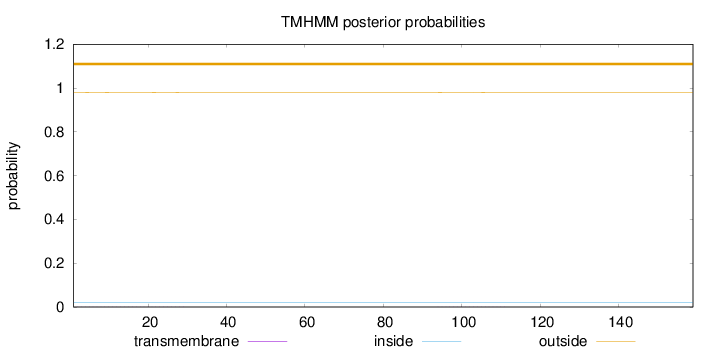
Topology
Length:
159
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01892
Exp number, first 60 AAs:
0.01476
Total prob of N-in:
0.02053
outside
1 - 159
Population Genetic Test Statistics
Pi
181.126395
Theta
177.110878
Tajima's D
0.183089
CLR
0.76976
CSRT
0.427678616069197
Interpretation
Uncertain
