Gene
KWMTBOMO12413
Pre Gene Modal
BGIBMGA012446
Annotation
nicotinic_acetylcholine_receptor_alpha_1_subunit_[Bombyx_mori]
Full name
Acetylcholine receptor subunit alpha-like 1
Alternative Name
Nicotinic acetylcholine receptor alpha 1
Location in the cell
PlasmaMembrane Reliability : 2.873
Sequence
CDS
ATGACAACGAACGTGTGGGTAGAACAGGAATGGAATGATTACAAGCTGAAATGGAACCCTGACGACTACGGTGGTGTGGACACGTTGCATGTGCCTTCAGAACACATATGGCTGCCAGATATCGTGCTTTACAACAACGCTGATGGCAACTACGAAGTGACGATAATGACGAAGGCGATCCTCCACCACGACGGCAAAGTGGTCTGGAAGCCGCCCGCCATTTACAAGTCCTTCTGCGAGATCGACGTTGAGTATTTTCCGTTCGACGAGCAAACCTGCTTCATGAAGTTCGGATCTTGGAGCTACGACGGGTACATGGTCGATCTGAGGCATTTAAAACAGTCGCCCGACTCGGACCACATTGGAATGGGAATAGATTTGTCAGAATATTACATCTCAGTTGAATGGGACATCATGCGGGTCCCAGCAACGAGAAACGAAAAGTTTTACTCTTGCTGTGAAGAACCATACCCGGATATTATCTTTAATATCACGCTCAGACGAAAAACTCTGTTCTACACAGTCAATCTTATTATTCCCTGCGTTGGAATTTCCTTCTTATCGGTTCTAGTATTCTATTTGCCTTCTGATTCAGGAGAGAAGATTTCACTTTGTATTTCAATCCTCCTATCTCTCACTGTGTTCTTCTTGCTGTTAGCAGAAATTATACCACCCACGTCACTTACGGTTCCGTTGCTTGGAAAATATTTGTTGTTCACAATGATGCTAGTTACGCTATCGGTAGTTGTCACTATCGTCGTACTCAACATAAATTTTAGGTCGCCCGTAACCCACAATATGGCCCCATGGGTGAGGAAAGTCTTTATAGATTTTTTACCCAAAATATTATTCATACAAAGGCCTGAAAAACCACCTGATGACGACGATGATAACGATAAACCCAGCGAGATACTCACGGATGTTTTTGGCCCAGACGATATGGATGGGAAATTCAAGGAGTGGGGTTGCGAGGAATACGATTTACCGGGGATGCCCCCTTCACCACCACCTCCTCCAGGAGGGGACGATGAGCTGTTCTCTCCACCTCCAGGATCCCCATGCCGTCTGGACCTTGACGATGGTAGCCCTTCTTTAGAAAAGCCGTACGTCAGAGAAATGGAAAAAACTATCGAAGGTTCTAGGTTTATAGCGCAACATGTCAAAAATAAGGATAAATTTGAAAGCGTTGAAGATGACTGGAAATACGTAGCGATGGTTTTAGACAGGATCTTTCTATTCCTGTTCACGATAGCATGCGTTCTCGGTACGGCGCTCATCATATTTAGAGCACCAACATTCTACGATAATACGAAACCTATCGACATTCTATACTCCAAAATAGCGAAGAAGAAGTTAGAGCTGCTCAAAATGGGCTCAGAAGGAGACCCCGGTCTCTGA
Protein
MTTNVWVEQEWNDYKLKWNPDDYGGVDTLHVPSEHIWLPDIVLYNNADGNYEVTIMTKAILHHDGKVVWKPPAIYKSFCEIDVEYFPFDEQTCFMKFGSWSYDGYMVDLRHLKQSPDSDHIGMGIDLSEYYISVEWDIMRVPATRNEKFYSCCEEPYPDIIFNITLRRKTLFYTVNLIIPCVGISFLSVLVFYLPSDSGEKISLCISILLSLTVFFLLLAEIIPPTSLTVPLLGKYLLFTMMLVTLSVVVTIVVLNINFRSPVTHNMAPWVRKVFIDFLPKILFIQRPEKPPDDDDDNDKPSEILTDVFGPDDMDGKFKEWGCEEYDLPGMPPSPPPPPGGDDELFSPPPGSPCRLDLDDGSPSLEKPYVREMEKTIEGSRFIAQHVKNKDKFESVEDDWKYVAMVLDRIFLFLFTIACVLGTALIIFRAPTFYDNTKPIDILYSKIAKKKLELLKMGSEGDPGL
Summary
Description
After binding acetylcholine, the AChR responds by an extensive change in conformation that affects all subunits and leads to opening of an ion-conducting channel across the plasma membrane.
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family. Acetylcholine receptor (TC 1.A.9.1) subfamily.
Belongs to the ligand-gated ion channel (TC 1.A.9) family. Acetylcholine receptor (TC 1.A.9.1) subfamily.
Keywords
Cell junction
Cell membrane
Complete proteome
Disulfide bond
Glycoprotein
Ion channel
Ion transport
Ligand-gated ion channel
Membrane
Polymorphism
Postsynaptic cell membrane
Receptor
Reference proteome
Signal
Synapse
Transmembrane
Transmembrane helix
Transport
Feature
chain Acetylcholine receptor subunit alpha-like 1
Uniprot
A8HTA7
A8CGK1
A0A2A4JXA4
A0A2H1WJZ5
A0A0H4U7T7
A0A212FPK4
+ More
A0A194R9X4 A0A194PWH0 A0A0L7LC91 H9JSD5 K7XPY0 B0WJI9 A0A0L0BP41 A0A182J166 A0A182P6P6 A0A182QWG6 W8C3D4 Q17DY3 Q7QD62 A0A182UUJ1 A0A1I8M4H7 A0A1I8P8R8 B4K9X9 B4LXX4 A0A3B0JM77 B4GE91 B4JFZ5 A0A1W4URW6 B5DWV2 A0A0B4KHE6 P09478 A0A0B4KGU3 E0WN28 B4PVU1 A0A1J1J1F2 B4NF15 B4HH23 B3P731 B3N1Q7 A0A0M4F5T1 B4QT66 A0A182WRP2 A0A182FNM7 A0A182HU36 A0A182XWD8 A0A182RN97 A0A1B0GDR4 A0A182KFD2 A0A0T6BCQ9 A0A2P8XKP1 R4I597 A0A1B6HT85 A8DIP3 B5ARH1 A0A1Y1KCU3 D3UA13 A0A067RI44 A0A232F011 E0VDF7 A0A2J7PSA3 A0A182MCM3 A0A348BSV1 A0A195DZA5 D2CVP1 K7IMG0 A0A195EVN3 N6UMS2 E2A4K4 A0A182NDV2 A0A1D8GS59 A0A3L8DMC0 A0A1W4WH52 U4TVN3 A0A182TCU4 A0A1D2MXL1 A0A1B0CSE8 A0A087UJR8 A0A1L4BJZ7 A0A336MI73 A0A1L4BJZ9 V9Z9T0 A0A088BD75 B7PBZ1 A0A1D2MEM2 A0A3B0KCA0 A0A1L4BJY5 A0A1B6CSI7 A0A2A4K147 O96632 O46133 Q8MUR0 W6EAG0 A0A0C9RZ38 A0A0N0U641 A0A0L7R4A8 A0A067RUF6 A0A2A3EKN1
A0A194R9X4 A0A194PWH0 A0A0L7LC91 H9JSD5 K7XPY0 B0WJI9 A0A0L0BP41 A0A182J166 A0A182P6P6 A0A182QWG6 W8C3D4 Q17DY3 Q7QD62 A0A182UUJ1 A0A1I8M4H7 A0A1I8P8R8 B4K9X9 B4LXX4 A0A3B0JM77 B4GE91 B4JFZ5 A0A1W4URW6 B5DWV2 A0A0B4KHE6 P09478 A0A0B4KGU3 E0WN28 B4PVU1 A0A1J1J1F2 B4NF15 B4HH23 B3P731 B3N1Q7 A0A0M4F5T1 B4QT66 A0A182WRP2 A0A182FNM7 A0A182HU36 A0A182XWD8 A0A182RN97 A0A1B0GDR4 A0A182KFD2 A0A0T6BCQ9 A0A2P8XKP1 R4I597 A0A1B6HT85 A8DIP3 B5ARH1 A0A1Y1KCU3 D3UA13 A0A067RI44 A0A232F011 E0VDF7 A0A2J7PSA3 A0A182MCM3 A0A348BSV1 A0A195DZA5 D2CVP1 K7IMG0 A0A195EVN3 N6UMS2 E2A4K4 A0A182NDV2 A0A1D8GS59 A0A3L8DMC0 A0A1W4WH52 U4TVN3 A0A182TCU4 A0A1D2MXL1 A0A1B0CSE8 A0A087UJR8 A0A1L4BJZ7 A0A336MI73 A0A1L4BJZ9 V9Z9T0 A0A088BD75 B7PBZ1 A0A1D2MEM2 A0A3B0KCA0 A0A1L4BJY5 A0A1B6CSI7 A0A2A4K147 O96632 O46133 Q8MUR0 W6EAG0 A0A0C9RZ38 A0A0N0U641 A0A0L7R4A8 A0A067RUF6 A0A2A3EKN1
Pubmed
17868469
22118469
26354079
26227816
19121390
23331538
+ More
26108605 24495485 17510324 12364791 14747013 15676276 17210077 25315136 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2840281 17550304 25244985 29403074 17880682 18362917 19820115 19533628 28004739 20087392 24845553 28648823 20566863 20075255 23537049 20798317 30249741 27289101 24291321 9660807 12752659 25701599
26108605 24495485 17510324 12364791 14747013 15676276 17210077 25315136 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2840281 17550304 25244985 29403074 17880682 18362917 19820115 19533628 28004739 20087392 24845553 28648823 20566863 20075255 23537049 20798317 30249741 27289101 24291321 9660807 12752659 25701599
EMBL
EU143792
ABV72683.1
EU082074
ABV45511.1
NWSH01000458
PCG76264.1
+ More
ODYU01009147 SOQ53369.1 KP711043 RSAL01000098 AKQ12739.1 RVE47607.1 AGBW02001736 OWR55681.1 KQ460473 KPJ14437.1 KQ459591 KPI97104.1 JTDY01001698 KOB73132.1 BABH01033782 KC134198 AFX68724.1 DS231960 EDS29185.1 JRES01001582 KNC21776.1 AXCN02001585 GAMC01010196 GAMC01010194 GAMC01010192 JAB96359.1 CH477290 EAT44618.1 AY705394 AAAB01008859 AAU12503.1 EAA08125.3 CH933806 EDW15627.2 CH940650 EDW66840.2 OUUW01000007 SPP83335.1 CH479182 EDW33926.1 CH916369 EDV92534.1 CM000070 EDY68186.2 AE014297 AGB96297.1 X07194 AGB96296.1 FR689743 CBX19379.1 CM000160 EDW98940.2 CVRI01000064 CRL05596.1 CH964251 EDW83390.2 CH480815 EDW43499.1 CH954182 EDV53851.1 CH902661 EDV34026.2 CP012526 ALC47300.1 CM000364 EDX14230.1 APCN01000604 CCAG010012008 CCAG010012009 CCAG010012010 CCAG010012011 CCAG010012012 CCAG010012013 CCAG010012014 CCAG010012015 CCAG010012016 CCAG010012017 LJIG01001953 KRT84987.1 PYGN01001836 PSN32586.1 JQ585634 KP725463 AFJ04793.1 AKV94620.1 GECU01029812 JAS77894.1 EF526080 KQ971312 ABS86902.1 EEZ99265.1 EU871527 ACG49260.1 GEZM01089799 JAV57435.1 FJ821429 ACY82683.1 KK852460 KDR23472.1 NNAY01001445 OXU23952.1 DS235075 EEB11413.1 NEVH01021935 PNF19215.1 AXCM01009820 LC389175 BBE49554.1 KQ980050 KYN18047.1 EU879965 ACJ64923.1 KQ981953 KYN32320.1 APGK01016960 KB739995 ENN82001.1 GL436710 EFN71606.1 KU557775 AOT81837.1 QOIP01000006 RLU21521.1 KB631626 ERL84827.1 LJIJ01000414 ODM97750.1 AJWK01025958 AJWK01025959 KK120139 KFM77607.1 KX021840 API81630.1 UFQT01000927 SSX28037.1 KX021828 API81618.1 KF695387 AHE40764.1 KF141793 AGV76068.1 ABJB010351222 DS680203 EEC04113.1 LJIJ01001600 ODM91301.1 SPP83336.1 KX021831 API81621.1 GEDC01020789 JAS16509.1 NWSH01000256 PCG77967.1 AF096879 AAD09809.1 AJ000391 CAA04053.1 AF514804 KJ939595 AAM51823.1 AJE70266.1 KF873589 AHJ11218.1 GBYB01013316 JAG83083.1 KQ435756 KOX75921.1 KQ414658 KOC65659.1 KDR23474.1 KZ288229 PBC31729.1
ODYU01009147 SOQ53369.1 KP711043 RSAL01000098 AKQ12739.1 RVE47607.1 AGBW02001736 OWR55681.1 KQ460473 KPJ14437.1 KQ459591 KPI97104.1 JTDY01001698 KOB73132.1 BABH01033782 KC134198 AFX68724.1 DS231960 EDS29185.1 JRES01001582 KNC21776.1 AXCN02001585 GAMC01010196 GAMC01010194 GAMC01010192 JAB96359.1 CH477290 EAT44618.1 AY705394 AAAB01008859 AAU12503.1 EAA08125.3 CH933806 EDW15627.2 CH940650 EDW66840.2 OUUW01000007 SPP83335.1 CH479182 EDW33926.1 CH916369 EDV92534.1 CM000070 EDY68186.2 AE014297 AGB96297.1 X07194 AGB96296.1 FR689743 CBX19379.1 CM000160 EDW98940.2 CVRI01000064 CRL05596.1 CH964251 EDW83390.2 CH480815 EDW43499.1 CH954182 EDV53851.1 CH902661 EDV34026.2 CP012526 ALC47300.1 CM000364 EDX14230.1 APCN01000604 CCAG010012008 CCAG010012009 CCAG010012010 CCAG010012011 CCAG010012012 CCAG010012013 CCAG010012014 CCAG010012015 CCAG010012016 CCAG010012017 LJIG01001953 KRT84987.1 PYGN01001836 PSN32586.1 JQ585634 KP725463 AFJ04793.1 AKV94620.1 GECU01029812 JAS77894.1 EF526080 KQ971312 ABS86902.1 EEZ99265.1 EU871527 ACG49260.1 GEZM01089799 JAV57435.1 FJ821429 ACY82683.1 KK852460 KDR23472.1 NNAY01001445 OXU23952.1 DS235075 EEB11413.1 NEVH01021935 PNF19215.1 AXCM01009820 LC389175 BBE49554.1 KQ980050 KYN18047.1 EU879965 ACJ64923.1 KQ981953 KYN32320.1 APGK01016960 KB739995 ENN82001.1 GL436710 EFN71606.1 KU557775 AOT81837.1 QOIP01000006 RLU21521.1 KB631626 ERL84827.1 LJIJ01000414 ODM97750.1 AJWK01025958 AJWK01025959 KK120139 KFM77607.1 KX021840 API81630.1 UFQT01000927 SSX28037.1 KX021828 API81618.1 KF695387 AHE40764.1 KF141793 AGV76068.1 ABJB010351222 DS680203 EEC04113.1 LJIJ01001600 ODM91301.1 SPP83336.1 KX021831 API81621.1 GEDC01020789 JAS16509.1 NWSH01000256 PCG77967.1 AF096879 AAD09809.1 AJ000391 CAA04053.1 AF514804 KJ939595 AAM51823.1 AJE70266.1 KF873589 AHJ11218.1 GBYB01013316 JAG83083.1 KQ435756 KOX75921.1 KQ414658 KOC65659.1 KDR23474.1 KZ288229 PBC31729.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000005204 UP000002320 UP000037069 UP000075880 UP000075885 UP000075886 UP000008820 UP000007062 UP000075903 UP000095301 UP000095300 UP000009192 UP000008792 UP000268350 UP000008744 UP000001070 UP000192221 UP000001819 UP000000803 UP000002282 UP000183832 UP000007798 UP000001292 UP000008711 UP000007801 UP000092553 UP000000304 UP000076407 UP000069272 UP000075840 UP000076408 UP000075900 UP000092444 UP000075881 UP000245037 UP000007266 UP000027135 UP000215335 UP000009046 UP000235965 UP000075883 UP000078492 UP000002358 UP000078541 UP000019118 UP000000311 UP000075884 UP000279307 UP000192223 UP000030742 UP000075902 UP000094527 UP000092461 UP000054359 UP000001555 UP000053105 UP000053825 UP000242457
UP000005204 UP000002320 UP000037069 UP000075880 UP000075885 UP000075886 UP000008820 UP000007062 UP000075903 UP000095301 UP000095300 UP000009192 UP000008792 UP000268350 UP000008744 UP000001070 UP000192221 UP000001819 UP000000803 UP000002282 UP000183832 UP000007798 UP000001292 UP000008711 UP000007801 UP000092553 UP000000304 UP000076407 UP000069272 UP000075840 UP000076408 UP000075900 UP000092444 UP000075881 UP000245037 UP000007266 UP000027135 UP000215335 UP000009046 UP000235965 UP000075883 UP000078492 UP000002358 UP000078541 UP000019118 UP000000311 UP000075884 UP000279307 UP000192223 UP000030742 UP000075902 UP000094527 UP000092461 UP000054359 UP000001555 UP000053105 UP000053825 UP000242457
Interpro
Gene 3D
ProteinModelPortal
A8HTA7
A8CGK1
A0A2A4JXA4
A0A2H1WJZ5
A0A0H4U7T7
A0A212FPK4
+ More
A0A194R9X4 A0A194PWH0 A0A0L7LC91 H9JSD5 K7XPY0 B0WJI9 A0A0L0BP41 A0A182J166 A0A182P6P6 A0A182QWG6 W8C3D4 Q17DY3 Q7QD62 A0A182UUJ1 A0A1I8M4H7 A0A1I8P8R8 B4K9X9 B4LXX4 A0A3B0JM77 B4GE91 B4JFZ5 A0A1W4URW6 B5DWV2 A0A0B4KHE6 P09478 A0A0B4KGU3 E0WN28 B4PVU1 A0A1J1J1F2 B4NF15 B4HH23 B3P731 B3N1Q7 A0A0M4F5T1 B4QT66 A0A182WRP2 A0A182FNM7 A0A182HU36 A0A182XWD8 A0A182RN97 A0A1B0GDR4 A0A182KFD2 A0A0T6BCQ9 A0A2P8XKP1 R4I597 A0A1B6HT85 A8DIP3 B5ARH1 A0A1Y1KCU3 D3UA13 A0A067RI44 A0A232F011 E0VDF7 A0A2J7PSA3 A0A182MCM3 A0A348BSV1 A0A195DZA5 D2CVP1 K7IMG0 A0A195EVN3 N6UMS2 E2A4K4 A0A182NDV2 A0A1D8GS59 A0A3L8DMC0 A0A1W4WH52 U4TVN3 A0A182TCU4 A0A1D2MXL1 A0A1B0CSE8 A0A087UJR8 A0A1L4BJZ7 A0A336MI73 A0A1L4BJZ9 V9Z9T0 A0A088BD75 B7PBZ1 A0A1D2MEM2 A0A3B0KCA0 A0A1L4BJY5 A0A1B6CSI7 A0A2A4K147 O96632 O46133 Q8MUR0 W6EAG0 A0A0C9RZ38 A0A0N0U641 A0A0L7R4A8 A0A067RUF6 A0A2A3EKN1
A0A194R9X4 A0A194PWH0 A0A0L7LC91 H9JSD5 K7XPY0 B0WJI9 A0A0L0BP41 A0A182J166 A0A182P6P6 A0A182QWG6 W8C3D4 Q17DY3 Q7QD62 A0A182UUJ1 A0A1I8M4H7 A0A1I8P8R8 B4K9X9 B4LXX4 A0A3B0JM77 B4GE91 B4JFZ5 A0A1W4URW6 B5DWV2 A0A0B4KHE6 P09478 A0A0B4KGU3 E0WN28 B4PVU1 A0A1J1J1F2 B4NF15 B4HH23 B3P731 B3N1Q7 A0A0M4F5T1 B4QT66 A0A182WRP2 A0A182FNM7 A0A182HU36 A0A182XWD8 A0A182RN97 A0A1B0GDR4 A0A182KFD2 A0A0T6BCQ9 A0A2P8XKP1 R4I597 A0A1B6HT85 A8DIP3 B5ARH1 A0A1Y1KCU3 D3UA13 A0A067RI44 A0A232F011 E0VDF7 A0A2J7PSA3 A0A182MCM3 A0A348BSV1 A0A195DZA5 D2CVP1 K7IMG0 A0A195EVN3 N6UMS2 E2A4K4 A0A182NDV2 A0A1D8GS59 A0A3L8DMC0 A0A1W4WH52 U4TVN3 A0A182TCU4 A0A1D2MXL1 A0A1B0CSE8 A0A087UJR8 A0A1L4BJZ7 A0A336MI73 A0A1L4BJZ9 V9Z9T0 A0A088BD75 B7PBZ1 A0A1D2MEM2 A0A3B0KCA0 A0A1L4BJY5 A0A1B6CSI7 A0A2A4K147 O96632 O46133 Q8MUR0 W6EAG0 A0A0C9RZ38 A0A0N0U641 A0A0L7R4A8 A0A067RUF6 A0A2A3EKN1
PDB
6CNK
E-value=1.65677e-76,
Score=728
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell junction
Synapse
Postsynaptic cell membrane
Cell membrane
Synapse
Postsynaptic cell membrane
Cell membrane
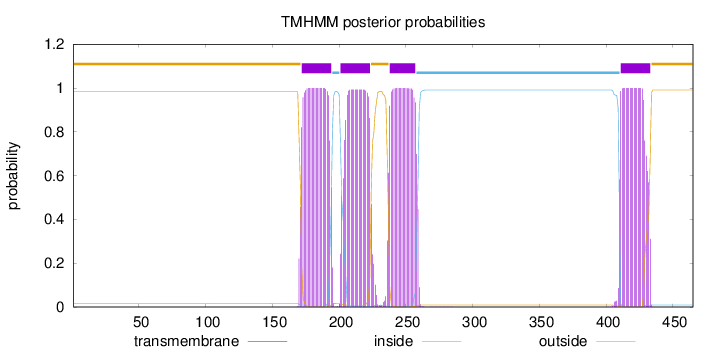
Length:
465
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
88.28762
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01571
outside
1 - 171
TMhelix
172 - 194
inside
195 - 200
TMhelix
201 - 223
outside
224 - 237
TMhelix
238 - 257
inside
258 - 410
TMhelix
411 - 433
outside
434 - 465
Population Genetic Test Statistics
Pi
182.386847
Theta
152.372474
Tajima's D
0.507941
CLR
0.668578
CSRT
0.517474126293685
Interpretation
Uncertain
