Gene
KWMTBOMO12409
Pre Gene Modal
BGIBMGA012422
Annotation
nicotinic_acetylcholine_receptor_subunit_alpha_2_precursor_[Bombyx_mori]
Full name
Acetylcholine receptor subunit alpha-like 2
+ More
Acetylcholine receptor subunit alpha-L1
Acetylcholine receptor subunit alpha-L1
Alternative Name
Nicotinic acetylcholine receptor alpha 2
Location in the cell
PlasmaMembrane Reliability : 4.46
Sequence
CDS
ATGTCTAAAGCTATTTTTGTCGTGTTTTTTTGTAGTTTTACTGTATGTTACGCGAATCCTGACGCGAAGAGGCTTTATGATGACTTACTGAGTAACTACAATAGACTGATTCGACCTGTTGATAAAAATAACAACACCGTACTAGTGAAGCTTGGGCTACGTCTATCCCAGTTGATTGATCTGAATTTGAAAGATCAAATATTAACGACTAACGTATGGTTGGAGCACGAATGGGAAGACCATAAGTTCAAATGGGATCCCTTGGAGTATGGTGGTGTAAGGGAGCTCTATGTGCCATCTGAACATATCTGGCTGCCAGATATAGTGCTTTATAACAATGCCGATGGTGAGTATGTTGTGACGACTATGACGAAAGCTGTGCTGCACCACAATGGAAAAGTGCTTTGGACTCCACCGGCCATCTTCAAGTCCTCCTGCGAGATCGACGTTCGATACTTCCCGTTCGATCAACAGACTTGCTTCTTGAAATTCGGATCTTGGAGCTATGATGGTGATCAGATCGACTTAAAACATATCAACCAAAAAAAGGGGGACATGGTCGAAGTTGGTATCGACCTTCGCGAGTACTACCCGTCAGTGGAATGGGATATTCTCGGAGTACCGGCTGAGAGACACGAGAGGTATTACCCCTGCTGTCAGGAACCGTATCCGGATATCTTCTTTAACATCACACTACGTAGGAAAACCTTATTCTATACTGTCAACCTAATCGTACCGTGTGTGGGGATTTCGTATCTGTCAGTTTTAGTATTTTATCTGCCCGCTGATTCCGGTGAAAAGATAGCCCTCAGCATTTCCATTCTTCTCTCACAAACTATGTTTTTTCTGCTCATTTCCGAAATTATTCCCTCGACTTCTCTAGCTTTACCTTTATTAGGAAAATATTTGCTATTCACCATGCTGCTCGTTGGTTTATCTGTTGTTATAACTATAATAATACTGAATGTACATTATCGAAAGCCTAGTACCCACAAAATGGCTCCATGGGTCAGAAAGTTCTTTATAACAAAACTTCCAAAGTTGCTGCTGATGAGAGTGCCGAAAGATTTGTTGAGAGATTTGGCCGCGCAGAAGATTGCCGGTAGGAGTATGAAGAACAAGAATAAATTCAAAGATGCGCTAGCGGCTGCCGATCAGACTCATTCTAATGCTTCAAGTCCAGACTCGCTCAGACATCACATGCCTGGAGGATGTAATGGACTGCATACTACAACAGCGACAAACAGATTTAGCGGTCTCGTTGGAGCATTAGGAAGCCTCGGTGCTGGATATAATGGTCTACCTTCCGTGATGTCAGGCCTCGATGATTCCTTGAGCGATGTGGTTCCGAGGAAGAAGTATCCGTTTGAATTAGAGAAAGCTATCCACAACGTCATGTTTATACAGCATCACATGCAGCGCCAAGATGAGTTTAATGCGGAAGATCAAGACTGGGGATTTGTGGCTATGGTATTGGATCGTCTGTTCCTGTGGATCTTCACCATAGCTTCCATAGTTGGTACTTTCGCAATCCTATGCGAAGCCCCGTCTTTGTACGACGACACGAAGCCGATTGACATGATGCTGTCTTCTGTGGCCCAGCAGCAATATCTACCTGTCGACAGTGGGGATTCATAA
Protein
MSKAIFVVFFCSFTVCYANPDAKRLYDDLLSNYNRLIRPVDKNNNTVLVKLGLRLSQLIDLNLKDQILTTNVWLEHEWEDHKFKWDPLEYGGVRELYVPSEHIWLPDIVLYNNADGEYVVTTMTKAVLHHNGKVLWTPPAIFKSSCEIDVRYFPFDQQTCFLKFGSWSYDGDQIDLKHINQKKGDMVEVGIDLREYYPSVEWDILGVPAERHERYYPCCQEPYPDIFFNITLRRKTLFYTVNLIVPCVGISYLSVLVFYLPADSGEKIALSISILLSQTMFFLLISEIIPSTSLALPLLGKYLLFTMLLVGLSVVITIIILNVHYRKPSTHKMAPWVRKFFITKLPKLLLMRVPKDLLRDLAAQKIAGRSMKNKNKFKDALAAADQTHSNASSPDSLRHHMPGGCNGLHTTTATNRFSGLVGALGSLGAGYNGLPSVMSGLDDSLSDVVPRKKYPFELEKAIHNVMFIQHHMQRQDEFNAEDQDWGFVAMVLDRLFLWIFTIASIVGTFAILCEAPSLYDDTKPIDMMLSSVAQQQYLPVDSGDS
Summary
Description
After binding acetylcholine, the AChR responds by an extensive change in conformation that affects all subunits and leads to opening of an ion-conducting channel across the plasma membrane.
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family. Acetylcholine receptor (TC 1.A.9.1) subfamily.
Belongs to the peptidase S1 family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family. Acetylcholine receptor (TC 1.A.9.1) subfamily.
Belongs to the peptidase S1 family.
Keywords
Cell junction
Cell membrane
Complete proteome
Disulfide bond
Glycoprotein
Ion channel
Ion transport
Ligand-gated ion channel
Membrane
Postsynaptic cell membrane
Receptor
Reference proteome
Signal
Synapse
Transmembrane
Transmembrane helix
Transport
Feature
chain Acetylcholine receptor subunit alpha-like 2
Uniprot
A8CGK2
A8HTB1
H9JSB1
O96631
A0A0H4TQB9
A0A0A7RQ69
+ More
A0A2H1W920 E0WN31 A0A154P4P0 A0A1A9ZCG6 A0A1A9X9A2 A0A1B0GDR8 A0A1A9VTK4 A0A0A1X0E1 Q17DY2 A0A195DYU8 F4WCW3 A0A2A3ESS5 B3N1R0 Q6QHT2 A0A0L7LL34 B4NF14 B4K9Y0 B4JFZ7 B4QT69 Q297D9 B4HH26 P17644 B4LXX5 B4PVT7 A0A195EVM9 B3P729 E2A4K9 A0A3B0KHZ9 A0A3L8DNB1 A0A182WQJ9 A0A2C9GPP7 Q7QD60 A0A034WMC6 A0A0M4EQK7 A1XI34 A0A0K8U8U8 A0A1I8NW57 A1XI33 A0A0L0BP06 K7WZZ6 A0A0C9QEW5 A0A1B0D962 A0A1J1J2B7 A0A310SK04 A0A2Z4N601 A0A348BSV2 A0A1W4V5Q5 A0A0K1U4H6 R4I3V1 A0A151IPU9 E9IZX9 D2CVN9 W6E9A5 E2C6B3 K7IMF9 A0A336M5I2 D3UA14 C3TS22 A8DIQ7 A0A067RKC1 C3TS21 C3TS18 P23414 E0VDF5 A0A1W4XPT8 A0A212EPA7 A0A1D8GS57 A0A026X1G6 A0A194R9Y0 A0A158NB31 A0A0N1PHV8 W8BLF1 A0A194PUL8 N6SS15 A0A0M8ZWT0 A0A0L7RAC4 A0A1B0BF23 A0A1B0CSE7 A0A084VZB4 A0A0U2GQV3 A0A2H8TRZ9 P91765 A0A224ANR7 B4GE90 A0A151I1N3 A0A1S3DA73 A2TM48 A0A3B0KB55 A0A226EXE3 A0A0J7KUP1 A0A2J7PSA1 A0A2P8XCL7 E9G087 A0A1D2MXT5
A0A2H1W920 E0WN31 A0A154P4P0 A0A1A9ZCG6 A0A1A9X9A2 A0A1B0GDR8 A0A1A9VTK4 A0A0A1X0E1 Q17DY2 A0A195DYU8 F4WCW3 A0A2A3ESS5 B3N1R0 Q6QHT2 A0A0L7LL34 B4NF14 B4K9Y0 B4JFZ7 B4QT69 Q297D9 B4HH26 P17644 B4LXX5 B4PVT7 A0A195EVM9 B3P729 E2A4K9 A0A3B0KHZ9 A0A3L8DNB1 A0A182WQJ9 A0A2C9GPP7 Q7QD60 A0A034WMC6 A0A0M4EQK7 A1XI34 A0A0K8U8U8 A0A1I8NW57 A1XI33 A0A0L0BP06 K7WZZ6 A0A0C9QEW5 A0A1B0D962 A0A1J1J2B7 A0A310SK04 A0A2Z4N601 A0A348BSV2 A0A1W4V5Q5 A0A0K1U4H6 R4I3V1 A0A151IPU9 E9IZX9 D2CVN9 W6E9A5 E2C6B3 K7IMF9 A0A336M5I2 D3UA14 C3TS22 A8DIQ7 A0A067RKC1 C3TS21 C3TS18 P23414 E0VDF5 A0A1W4XPT8 A0A212EPA7 A0A1D8GS57 A0A026X1G6 A0A194R9Y0 A0A158NB31 A0A0N1PHV8 W8BLF1 A0A194PUL8 N6SS15 A0A0M8ZWT0 A0A0L7RAC4 A0A1B0BF23 A0A1B0CSE7 A0A084VZB4 A0A0U2GQV3 A0A2H8TRZ9 P91765 A0A224ANR7 B4GE90 A0A151I1N3 A0A1S3DA73 A2TM48 A0A3B0KB55 A0A226EXE3 A0A0J7KUP1 A0A2J7PSA1 A0A2P8XCL7 E9G087 A0A1D2MXT5
Pubmed
17868469
19121390
25504620
25830018
17510324
21719571
+ More
17994087 18057021 15656979 26227816 15632085 2117557 1697262 2114015 10731132 12537572 12537569 17550304 20798317 30249741 12364791 14747013 15676276 17210077 25348373 17167752 25315136 26108605 23331538 21282665 25701599 20075255 20087392 19320762 17880682 18362917 19820115 24845553 1702381 20566863 22118469 24508170 26354079 21347285 24495485 23537049 24438588 29403074 21292972 27289101
17994087 18057021 15656979 26227816 15632085 2117557 1697262 2114015 10731132 12537572 12537569 17550304 20798317 30249741 12364791 14747013 15676276 17210077 25348373 17167752 25315136 26108605 23331538 21282665 25701599 20075255 20087392 19320762 17880682 18362917 19820115 24845553 1702381 20566863 22118469 24508170 26354079 21347285 24495485 23537049 24438588 29403074 21292972 27289101
EMBL
EU082075
ABV45512.1
EU143793
ABV72684.1
BABH01033749
BABH01033750
+ More
BABH01033751 AF096878 NWSH01000941 AAD09808.1 PCG73442.1 KP711044 RSAL01000098 AKQ12740.1 RVE47610.1 KP101241 AJA39817.1 ODYU01006800 SOQ48984.1 FR689746 CBX19382.1 KQ434804 KZC06080.1 CCAG010012019 GBXI01009513 JAD04779.1 CH477290 EAT44619.1 KQ980050 KYN18043.1 GL888074 EGI67944.1 KZ288193 PBC34179.1 CH902661 EDV34029.1 KPU74901.1 AY540846 KJ939589 AAS48080.1 AJE70260.1 JTDY01000730 KOB76074.1 CH964251 EDW83389.1 CH933806 EDW15628.1 CH916369 EDV92536.1 CM000364 EDX14233.1 CM000070 EAL28267.2 CH480815 EDW43502.1 X52274 X53583 AE014297 AY058446 CH940650 EDW66841.1 CM000160 EDW98936.1 KQ981953 KYN32315.1 CH954182 EDV53849.1 GL436710 EFN71611.1 OUUW01000007 SPP83338.1 QOIP01000006 RLU21713.1 APCN01000604 AY705395 AAAB01008859 AAU12504.1 EAA08142.3 GAKP01004009 JAC54943.1 CP012526 ALC47301.1 DQ372063 ABD37618.1 GDHF01029569 GDHF01021142 GDHF01017588 GDHF01014388 JAI22745.1 JAI31172.1 JAI34726.1 JAI37926.1 DQ372062 ABD37617.1 JRES01001582 KNC21741.1 KC134201 AFX68727.1 GBYB01001869 JAG71636.1 AJVK01004243 AJVK01004244 CVRI01000064 CRL05582.1 KQ762207 OAD56063.1 MF612144 AWX65632.1 LC389176 BBE49555.1 KP725464 AKV94621.1 JQ585635 AFJ04794.1 KQ976812 KYN08175.1 GL767291 EFZ13791.1 EU879963 ACJ64921.1 KF873580 AHJ11209.1 GL453058 EFN76512.1 UFQT01000425 SSX24213.1 FJ821430 ACY82684.1 EU930052 EU930053 EU930054 ACM09847.1 EF526081 KQ971372 ABS86903.1 EFA10793.1 KK852460 KDR23473.1 EU930051 ACM09846.1 EU926748 ACL00341.1 X55439 DS235075 EEB11411.1 AGBW02013495 OWR43335.1 KU557776 AOT81838.1 KK107039 EZA61861.1 KQ460473 KPJ14442.1 ADTU01010787 LADI01003211 KPJ20580.1 GAMC01008708 GAMC01008707 JAB97848.1 KQ459591 KPI97101.1 APGK01059131 APGK01059132 KB741292 ENN70434.1 KQ435845 KOX71203.1 KQ414618 KOC67810.1 JXJN01013271 JXJN01013272 AJWK01025954 AJWK01025955 ATLV01018685 ATLV01018686 ATLV01018687 ATLV01018688 ATLV01018689 ATLV01018690 ATLV01018691 ATLV01018692 ATLV01018693 KE525245 KFB43308.1 KT328067 ALB27745.1 GFXV01004846 MBW16651.1 X81887 CAA57476.1 LC215866 BBA21161.1 CH479182 EDW33925.1 KQ976572 KYM80413.1 EF363700 ABM97499.1 SPP83339.1 LNIX01000001 OXA61860.1 LBMM01003055 KMQ94021.1 NEVH01021935 PNF19212.1 PYGN01003029 PSN29730.1 GL732528 EFX86859.1 LJIJ01000414 ODM97751.1
BABH01033751 AF096878 NWSH01000941 AAD09808.1 PCG73442.1 KP711044 RSAL01000098 AKQ12740.1 RVE47610.1 KP101241 AJA39817.1 ODYU01006800 SOQ48984.1 FR689746 CBX19382.1 KQ434804 KZC06080.1 CCAG010012019 GBXI01009513 JAD04779.1 CH477290 EAT44619.1 KQ980050 KYN18043.1 GL888074 EGI67944.1 KZ288193 PBC34179.1 CH902661 EDV34029.1 KPU74901.1 AY540846 KJ939589 AAS48080.1 AJE70260.1 JTDY01000730 KOB76074.1 CH964251 EDW83389.1 CH933806 EDW15628.1 CH916369 EDV92536.1 CM000364 EDX14233.1 CM000070 EAL28267.2 CH480815 EDW43502.1 X52274 X53583 AE014297 AY058446 CH940650 EDW66841.1 CM000160 EDW98936.1 KQ981953 KYN32315.1 CH954182 EDV53849.1 GL436710 EFN71611.1 OUUW01000007 SPP83338.1 QOIP01000006 RLU21713.1 APCN01000604 AY705395 AAAB01008859 AAU12504.1 EAA08142.3 GAKP01004009 JAC54943.1 CP012526 ALC47301.1 DQ372063 ABD37618.1 GDHF01029569 GDHF01021142 GDHF01017588 GDHF01014388 JAI22745.1 JAI31172.1 JAI34726.1 JAI37926.1 DQ372062 ABD37617.1 JRES01001582 KNC21741.1 KC134201 AFX68727.1 GBYB01001869 JAG71636.1 AJVK01004243 AJVK01004244 CVRI01000064 CRL05582.1 KQ762207 OAD56063.1 MF612144 AWX65632.1 LC389176 BBE49555.1 KP725464 AKV94621.1 JQ585635 AFJ04794.1 KQ976812 KYN08175.1 GL767291 EFZ13791.1 EU879963 ACJ64921.1 KF873580 AHJ11209.1 GL453058 EFN76512.1 UFQT01000425 SSX24213.1 FJ821430 ACY82684.1 EU930052 EU930053 EU930054 ACM09847.1 EF526081 KQ971372 ABS86903.1 EFA10793.1 KK852460 KDR23473.1 EU930051 ACM09846.1 EU926748 ACL00341.1 X55439 DS235075 EEB11411.1 AGBW02013495 OWR43335.1 KU557776 AOT81838.1 KK107039 EZA61861.1 KQ460473 KPJ14442.1 ADTU01010787 LADI01003211 KPJ20580.1 GAMC01008708 GAMC01008707 JAB97848.1 KQ459591 KPI97101.1 APGK01059131 APGK01059132 KB741292 ENN70434.1 KQ435845 KOX71203.1 KQ414618 KOC67810.1 JXJN01013271 JXJN01013272 AJWK01025954 AJWK01025955 ATLV01018685 ATLV01018686 ATLV01018687 ATLV01018688 ATLV01018689 ATLV01018690 ATLV01018691 ATLV01018692 ATLV01018693 KE525245 KFB43308.1 KT328067 ALB27745.1 GFXV01004846 MBW16651.1 X81887 CAA57476.1 LC215866 BBA21161.1 CH479182 EDW33925.1 KQ976572 KYM80413.1 EF363700 ABM97499.1 SPP83339.1 LNIX01000001 OXA61860.1 LBMM01003055 KMQ94021.1 NEVH01021935 PNF19212.1 PYGN01003029 PSN29730.1 GL732528 EFX86859.1 LJIJ01000414 ODM97751.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000076502
UP000092445
UP000092443
+ More
UP000092444 UP000078200 UP000008820 UP000078492 UP000007755 UP000242457 UP000007801 UP000005203 UP000037510 UP000007798 UP000009192 UP000001070 UP000000304 UP000001819 UP000001292 UP000000803 UP000008792 UP000002282 UP000078541 UP000008711 UP000000311 UP000268350 UP000279307 UP000075920 UP000075840 UP000007062 UP000092553 UP000095301 UP000095300 UP000037069 UP000092462 UP000183832 UP000192221 UP000078542 UP000008237 UP000002358 UP000007266 UP000027135 UP000009046 UP000192223 UP000007151 UP000053097 UP000053240 UP000005205 UP000053268 UP000019118 UP000053105 UP000053825 UP000092460 UP000092461 UP000030765 UP000008744 UP000078540 UP000079169 UP000198287 UP000036403 UP000235965 UP000245037 UP000000305 UP000094527
UP000092444 UP000078200 UP000008820 UP000078492 UP000007755 UP000242457 UP000007801 UP000005203 UP000037510 UP000007798 UP000009192 UP000001070 UP000000304 UP000001819 UP000001292 UP000000803 UP000008792 UP000002282 UP000078541 UP000008711 UP000000311 UP000268350 UP000279307 UP000075920 UP000075840 UP000007062 UP000092553 UP000095301 UP000095300 UP000037069 UP000092462 UP000183832 UP000192221 UP000078542 UP000008237 UP000002358 UP000007266 UP000027135 UP000009046 UP000192223 UP000007151 UP000053097 UP000053240 UP000005205 UP000053268 UP000019118 UP000053105 UP000053825 UP000092460 UP000092461 UP000030765 UP000008744 UP000078540 UP000079169 UP000198287 UP000036403 UP000235965 UP000245037 UP000000305 UP000094527
Interpro
IPR006202
Neur_chan_lig-bd
+ More
IPR006029 Neurotrans-gated_channel_TM
IPR036734 Neur_chan_lig-bd_sf
IPR002394 Nicotinic_acetylcholine_rcpt
IPR036719 Neuro-gated_channel_TM_sf
IPR018000 Neurotransmitter_ion_chnl_CS
IPR006201 Neur_channel
IPR033116 TRYPSIN_SER
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR032135 DUF4817
IPR006029 Neurotrans-gated_channel_TM
IPR036734 Neur_chan_lig-bd_sf
IPR002394 Nicotinic_acetylcholine_rcpt
IPR036719 Neuro-gated_channel_TM_sf
IPR018000 Neurotransmitter_ion_chnl_CS
IPR006201 Neur_channel
IPR033116 TRYPSIN_SER
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR032135 DUF4817
Gene 3D
CDD
ProteinModelPortal
A8CGK2
A8HTB1
H9JSB1
O96631
A0A0H4TQB9
A0A0A7RQ69
+ More
A0A2H1W920 E0WN31 A0A154P4P0 A0A1A9ZCG6 A0A1A9X9A2 A0A1B0GDR8 A0A1A9VTK4 A0A0A1X0E1 Q17DY2 A0A195DYU8 F4WCW3 A0A2A3ESS5 B3N1R0 Q6QHT2 A0A0L7LL34 B4NF14 B4K9Y0 B4JFZ7 B4QT69 Q297D9 B4HH26 P17644 B4LXX5 B4PVT7 A0A195EVM9 B3P729 E2A4K9 A0A3B0KHZ9 A0A3L8DNB1 A0A182WQJ9 A0A2C9GPP7 Q7QD60 A0A034WMC6 A0A0M4EQK7 A1XI34 A0A0K8U8U8 A0A1I8NW57 A1XI33 A0A0L0BP06 K7WZZ6 A0A0C9QEW5 A0A1B0D962 A0A1J1J2B7 A0A310SK04 A0A2Z4N601 A0A348BSV2 A0A1W4V5Q5 A0A0K1U4H6 R4I3V1 A0A151IPU9 E9IZX9 D2CVN9 W6E9A5 E2C6B3 K7IMF9 A0A336M5I2 D3UA14 C3TS22 A8DIQ7 A0A067RKC1 C3TS21 C3TS18 P23414 E0VDF5 A0A1W4XPT8 A0A212EPA7 A0A1D8GS57 A0A026X1G6 A0A194R9Y0 A0A158NB31 A0A0N1PHV8 W8BLF1 A0A194PUL8 N6SS15 A0A0M8ZWT0 A0A0L7RAC4 A0A1B0BF23 A0A1B0CSE7 A0A084VZB4 A0A0U2GQV3 A0A2H8TRZ9 P91765 A0A224ANR7 B4GE90 A0A151I1N3 A0A1S3DA73 A2TM48 A0A3B0KB55 A0A226EXE3 A0A0J7KUP1 A0A2J7PSA1 A0A2P8XCL7 E9G087 A0A1D2MXT5
A0A2H1W920 E0WN31 A0A154P4P0 A0A1A9ZCG6 A0A1A9X9A2 A0A1B0GDR8 A0A1A9VTK4 A0A0A1X0E1 Q17DY2 A0A195DYU8 F4WCW3 A0A2A3ESS5 B3N1R0 Q6QHT2 A0A0L7LL34 B4NF14 B4K9Y0 B4JFZ7 B4QT69 Q297D9 B4HH26 P17644 B4LXX5 B4PVT7 A0A195EVM9 B3P729 E2A4K9 A0A3B0KHZ9 A0A3L8DNB1 A0A182WQJ9 A0A2C9GPP7 Q7QD60 A0A034WMC6 A0A0M4EQK7 A1XI34 A0A0K8U8U8 A0A1I8NW57 A1XI33 A0A0L0BP06 K7WZZ6 A0A0C9QEW5 A0A1B0D962 A0A1J1J2B7 A0A310SK04 A0A2Z4N601 A0A348BSV2 A0A1W4V5Q5 A0A0K1U4H6 R4I3V1 A0A151IPU9 E9IZX9 D2CVN9 W6E9A5 E2C6B3 K7IMF9 A0A336M5I2 D3UA14 C3TS22 A8DIQ7 A0A067RKC1 C3TS21 C3TS18 P23414 E0VDF5 A0A1W4XPT8 A0A212EPA7 A0A1D8GS57 A0A026X1G6 A0A194R9Y0 A0A158NB31 A0A0N1PHV8 W8BLF1 A0A194PUL8 N6SS15 A0A0M8ZWT0 A0A0L7RAC4 A0A1B0BF23 A0A1B0CSE7 A0A084VZB4 A0A0U2GQV3 A0A2H8TRZ9 P91765 A0A224ANR7 B4GE90 A0A151I1N3 A0A1S3DA73 A2TM48 A0A3B0KB55 A0A226EXE3 A0A0J7KUP1 A0A2J7PSA1 A0A2P8XCL7 E9G087 A0A1D2MXT5
PDB
6CNK
E-value=1.38397e-102,
Score=954
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell junction
Synapse
Postsynaptic cell membrane
Cell membrane
Synapse
Postsynaptic cell membrane
Cell membrane
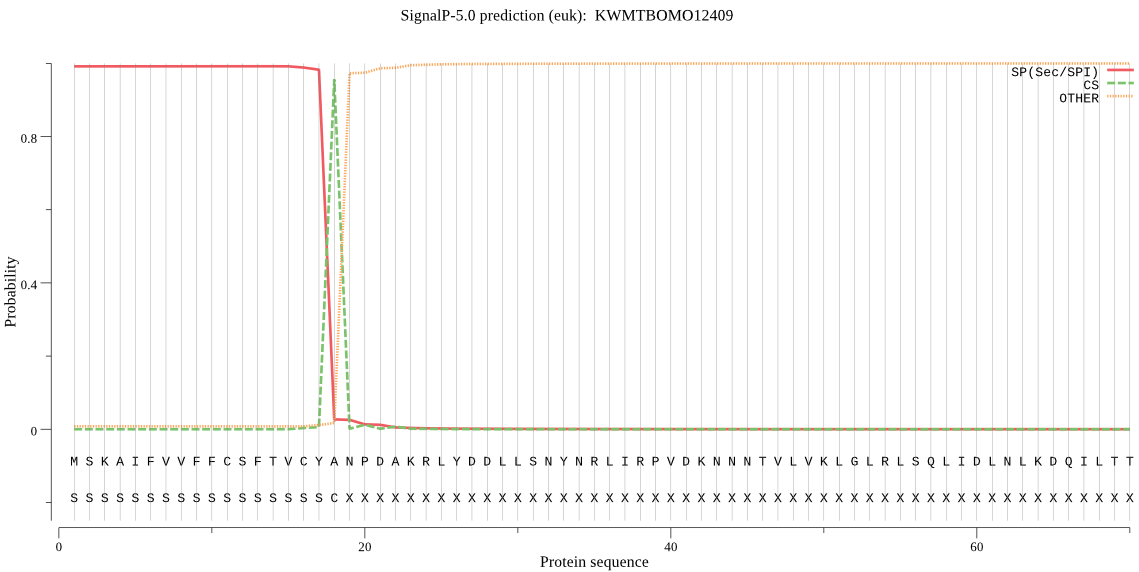
SignalP
Position: 1 - 18,
Likelihood: 0.991388
Length:
545
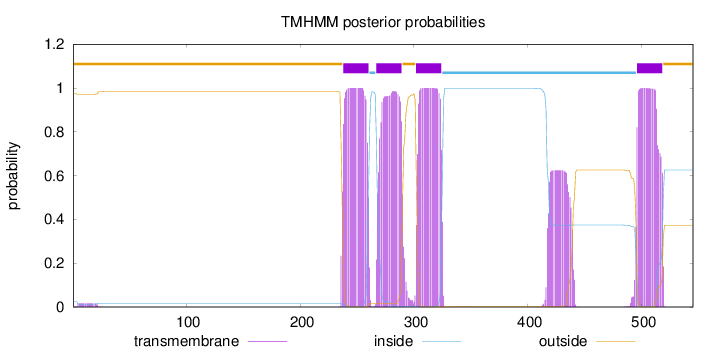
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
102.47751
Exp number, first 60 AAs:
0.29327
Total prob of N-in:
0.02452
outside
1 - 237
TMhelix
238 - 260
inside
261 - 266
TMhelix
267 - 289
outside
290 - 301
TMhelix
302 - 324
inside
325 - 495
TMhelix
496 - 518
outside
519 - 545
Population Genetic Test Statistics
Pi
181.728587
Theta
158.081395
Tajima's D
0.387371
CLR
0.173877
CSRT
0.48297585120744
Interpretation
Uncertain
