Gene
KWMTBOMO12408 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012423
Annotation
PREDICTED:_serine_protease_snake-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.099 Nuclear Reliability : 1.241
Sequence
CDS
ATGTATTTCTGCTGTTTATTCATAATTTTATTTGCCGATTCAGTTCACAACACTGTGCCCACGTCTCCCCGAGTCGGAGGCAACCGTGAGAAAATAATAAAACGAAGAGCCATAAAAGAAGACGATTGGATATGGGGACCCGTTGATCGCATTGTAAACAGCAAAAACGAAAATTCATCTTTTCCGTTAGGTACATGGTCTCCATTGGAAAACGACGACAAACCTTGGCTACACCGCATCAGATTTCCACCTTTTGCATACAGTTCCGCCCCTGATGATCTTATATTAATCCAACCAATAATCCAAGTTAACTCAACGAGAAAAATCAAAGTGATACCTCAATTACAAAATTATAATTTACCACCTATAAATAGTGAGGGAAAACAAAACATTCTATTGAAGTTTGTGATAGAATATGATAAAGAAGAAAACGAAATTACGTTAGAAGCAAGGACTGCTGTGAATGAAACAAATGATATTCCTTTACAGAGTACAGACAAAATACAAAGCTCAGAATTCAATAAAACAAACAATAATGGTCTTCTATCGGCATCTATTAAAGAACCAAATAGTGAACTAATAAGCAATGCGGTTTTAAACAATCAAACTAAAATCAAAGAAAACATAAAAGAAGAAAACATTACGACCAGAATCAATACCGAAGACGAAGAACCATCAATACAGACTTTAAACTCTTTCCAAATATTGTATTCTGAATTAAATTTTCCAAAGAAGAATAATAGAGTCATTGTAAAACCAGATCCTCCGTGTAAAGAATTCGAACTGCCTATATTTAGAAAAGGCTCGCGTATCAGTGAAACCAAATGCCGAGAACAAATTTGGAAATTAAACGCAATGGAGCGAATAGACACTGAAGATTACATCTGTCATTACAATCCGGATGCTGCAGTCGGAGGAAGAGATACATTCCCTGGTGAATTTCCACATATGGGCGCTGTTGGATGGTCCCCGTGGGATAAAAATTCGGATTACGTATTCAAATGCGGAAGCTCCCTAATCAGTCCATTTTTCTTACTGACTGCTGCTCACTGTTCAGAAGCACCGCTGAAGGATAATCCTGATATATCAGATCCGATCCCAAAGATTGTGAGACTGAGTTACAAAAATCTCTACGCTAAAAAACCAAGAGATAACTATCAGGACTCCAAGATAAAAGCAATTTACGTTCATCCCGATTACAAGTCTCCGACCAAATATAACGATGTAGCTTTGTTAGAATTAGAGTGGGGGATTAATTACTCATACTTTGTACAACCTGCGTGTCTATGGACGAAAGAAGACATAACGGAATTGGAATCTTTCGCCACCGTCACCGGCTGGGGAGTTTTACAAGAAGGTAGCAGGAACATCTCAGCCGAATTACAAGCAGCGGTAGTGGACTTTATAGATTCAGATGAATGCGAAACACTATTGCAACCTTGGTGCAGCAGGAATTGGTGTGGACTTGAAGAAACACAGCTATGTGCTGGCGTACTAGCAGGAGGCGTGGATGCATGTCAGGGTGATTCTGGAGGACCCCTCCAAGTGAAGATTTCCTTGAACTTCAGGTCAACGTACAATCTCTATTACATTGTCGGTGTTACGTCATTTGGTATTGGTTGTGCACAACCTAACATACCAGGAGTTTACACTAGGGTATCATCCTTTATAGACTGGATAGAAGAAAAAGTTTGGCCTGTCAACAAGAATGTCACTACGTAA
Protein
MYFCCLFIILFADSVHNTVPTSPRVGGNREKIIKRRAIKEDDWIWGPVDRIVNSKNENSSFPLGTWSPLENDDKPWLHRIRFPPFAYSSAPDDLILIQPIIQVNSTRKIKVIPQLQNYNLPPINSEGKQNILLKFVIEYDKEENEITLEARTAVNETNDIPLQSTDKIQSSEFNKTNNNGLLSASIKEPNSELISNAVLNNQTKIKENIKEENITTRINTEDEEPSIQTLNSFQILYSELNFPKKNNRVIVKPDPPCKEFELPIFRKGSRISETKCREQIWKLNAMERIDTEDYICHYNPDAAVGGRDTFPGEFPHMGAVGWSPWDKNSDYVFKCGSSLISPFFLLTAAHCSEAPLKDNPDISDPIPKIVRLSYKNLYAKKPRDNYQDSKIKAIYVHPDYKSPTKYNDVALLELEWGINYSYFVQPACLWTKEDITELESFATVTGWGVLQEGSRNISAELQAAVVDFIDSDECETLLQPWCSRNWCGLEETQLCAGVLAGGVDACQGDSGGPLQVKISLNFRSTYNLYYIVGVTSFGIGCAQPNIPGVYTRVSSFIDWIEEKVWPVNKNVTT
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
Proteomes
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
PDB
6O1G
E-value=9.6563e-32,
Score=343
Ontologies
GO
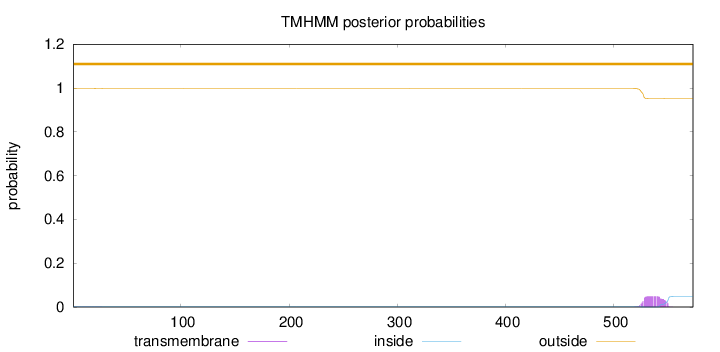
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.945509
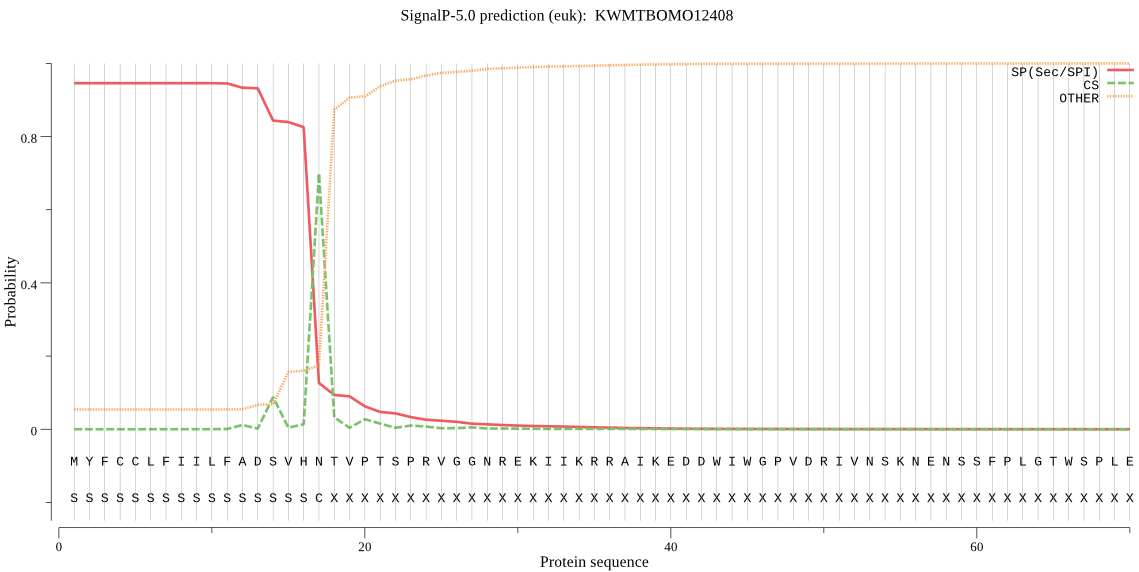
Length:
573
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.06601
Exp number, first 60 AAs:
0.01862
Total prob of N-in:
0.00210
outside
1 - 573
Population Genetic Test Statistics
Pi
190.427814
Theta
13.124889
Tajima's D
0.287559
CLR
0.079704
CSRT
0.461226938653067
Interpretation
Uncertain
