Gene
KWMTBOMO12405 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012427
Annotation
serine_protease_7_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.652
Sequence
CDS
ATGGCGGTTACATCGTTATTGAGACTGGAAGTATTGTTTGCAATAACCTTTTGCATCGTGGCCGAAGTGTTAATGCACCCAGAAAACGACGCACTGCAAGGTTCTAGTGATAACAACTCTGATGGAAATAAACGGCCAGAAGAAATATGGTCTTGGGGCTATGTGGACAATACAAAATCCAACAACCAGTCTGCAACCACCGAAGAACCTGAAGTGTGCAAACCGTATAATCCAACAGTACCCAACTTCGAGAAACCTGGAAGAAGAATCAGTGAAGTCAAATGCTATGAATATCTATGGAATATAAATTTCAGAGAAGAGCGTGATCGAAGATTGGACAAATGCTTTAAGCTGCATAGTAATGTCCAGCCATCATTTGCTATTGGGGGACGTAACACATTACCTGGCGAATTTCCACACATGGGTGCCATAGGCTGGCAGGCAGTCGTTGGATCCTGGATATTCAAATGTGGTGGATCGTTGATTAGCAATAAATTTATACTGACTGCGGCTCATTGTACGAGCTTCTCCCTGAAAGACACTACCATCGCTGATCCTATTCCTAAGATCGTCAGACTCGGCGACAAATATATTTTAGACAAGGAAGTAAACGATGGCATAATCCCTGAAGATAGAGAGATCGTAAATATCATTAAACATCCATCCTACAATCCTCCGAAGAAATACTACGACATTGCCCTCATGGAGTTAGACAAAGATGTCTTCTTCAGCAAGTACGTGCAGCCGGCATGCCTTTGGCCTCACTTCGATCTCAGCAGTTTGGGGAAAAAAGCTAGTGCGACAGGATGGGGAGTTGTCGATGCAAGAAGCACTGACATATCGCCCGAACTGCAAGCTATCGTAATAGATTTGATAGATACCCCGCAATGTCAGCAATTGCTCGAGACTTCGTGCAACAGACACTGGTGCGGTGTCGAAGATCATCAGCTGTGCGCTGGGAAGTTGGCTGGGGGAGTGGATGCATGTCAGGGTGACTCTGGAGGTCCTCTCCAAGTTGAAATATCTCTTCCTACATCGTCTCAAGGCAAGATATATTGTATAATCGGCGTGACGTCATTCGGGATCGGCTGTGCTCTCCCTGAACTGCCCGGGATATACACGAGAGTGTCGAGCTTCATTGATTGGATCGAACAGAATGTTTGGCCTACAGTCCACCAAAGTGGTACAGCTTAG
Protein
MAVTSLLRLEVLFAITFCIVAEVLMHPENDALQGSSDNNSDGNKRPEEIWSWGYVDNTKSNNQSATTEEPEVCKPYNPTVPNFEKPGRRISEVKCYEYLWNINFREERDRRLDKCFKLHSNVQPSFAIGGRNTLPGEFPHMGAIGWQAVVGSWIFKCGGSLISNKFILTAAHCTSFSLKDTTIADPIPKIVRLGDKYILDKEVNDGIIPEDREIVNIIKHPSYNPPKKYYDIALMELDKDVFFSKYVQPACLWPHFDLSSLGKKASATGWGVVDARSTDISPELQAIVIDLIDTPQCQQLLETSCNRHWCGVEDHQLCAGKLAGGVDACQGDSGGPLQVEISLPTSSQGKIYCIIGVTSFGIGCALPELPGIYTRVSSFIDWIEQNVWPTVHQSGTA
Summary
Similarity
Belongs to the peptidase S1 family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
H9JSB6
Q1HPQ6
Q5MPC5
A0A2H1W7F8
I4DLF7
A0A3S2NHC8
+ More
A0A437BB50 A0A212F1T1 A0A212EPD4 A0A2H1WUS1 A0A3S2LZB7 A0A2A4JE90 A0A194RAB5 A0A437BB20 A0A2A4JFQ7 A0A212EHP9 A0A194R9C5 I4DLF6 A0A437BB81 A0A194PUL8 A0A437BWN7 A0A194PLU8 A0A2A4JQ23 A0A2W1B5H2 I4DMI4 A0A2H1VIS4 G9F9H9 A0A437BB41 I4DQ94 A0A212F1E7 A0A194RM65 A0A0N0PBX6 E0V917 A0A084VW05 Q9XY63 A0A1J1J158 A0A067RX29 A0A0N1PHM7 A0A1S4FI15 Q16ZZ9 A0A1Y1KAU3 A0A1Y9HED1 A0A182JGW6 A0A023EQX6 A0A2J7RDN5 A0A2C9GS29 A0A023EPR9 F5HLC4 A0A084W6V6 A0A2J7RDN0 Q170A0 A0A182ID15 A0A182VFE5 A0A182X8T2 J9HYX9 X1WIF2 A0A182LU74 A0A182U5J8 A0A1J1J052 A0A154P3Y1
A0A437BB50 A0A212F1T1 A0A212EPD4 A0A2H1WUS1 A0A3S2LZB7 A0A2A4JE90 A0A194RAB5 A0A437BB20 A0A2A4JFQ7 A0A212EHP9 A0A194R9C5 I4DLF6 A0A437BB81 A0A194PUL8 A0A437BWN7 A0A194PLU8 A0A2A4JQ23 A0A2W1B5H2 I4DMI4 A0A2H1VIS4 G9F9H9 A0A437BB41 I4DQ94 A0A212F1E7 A0A194RM65 A0A0N0PBX6 E0V917 A0A084VW05 Q9XY63 A0A1J1J158 A0A067RX29 A0A0N1PHM7 A0A1S4FI15 Q16ZZ9 A0A1Y1KAU3 A0A1Y9HED1 A0A182JGW6 A0A023EQX6 A0A2J7RDN5 A0A2C9GS29 A0A023EPR9 F5HLC4 A0A084W6V6 A0A2J7RDN0 Q170A0 A0A182ID15 A0A182VFE5 A0A182X8T2 J9HYX9 X1WIF2 A0A182LU74 A0A182U5J8 A0A1J1J052 A0A154P3Y1
Pubmed
EMBL
BABH01033742
DQ443346
ABF51435.1
AY672785
AAV91007.1
ODYU01006800
+ More
SOQ48987.1 AK402125 BAM18747.1 RSAL01000098 RVE47611.1 RVE47614.1 AGBW02010816 OWR47692.1 AGBW02013495 OWR43336.1 ODYU01011151 SOQ56716.1 RVE47596.1 NWSH01001694 PCG70405.1 KQ460473 KPJ14444.1 RVE47617.1 PCG70404.1 AGBW02014853 OWR40990.1 KPJ14443.1 AK402124 BAM18746.1 RVE47613.1 KQ459591 KPI97101.1 RSAL01000002 RVE54897.1 KQ459600 KPI94302.1 NWSH01000865 PCG73808.1 KZ150313 PZC71449.1 AK402502 BAM19124.1 ODYU01002797 SOQ40740.1 JN033724 AEW46877.1 RVE47618.1 AK403997 BAM20084.1 AGBW02010907 OWR47533.1 KQ459995 KPJ18524.1 KQ460736 KPJ12593.1 DS234988 EEB09873.1 ATLV01017365 KE525164 KFB42149.1 AF053921 AAD21841.1 CVRI01000064 CRL05174.1 KK852411 KDR24454.1 KQ458725 KPJ05344.1 CH477477 EAT40291.1 GEZM01092351 JAV56586.1 GAPW01002075 JAC11523.1 NEVH01005287 PNF38945.1 APCN01003246 GAPW01002076 JAC11522.1 AAAB01008888 EGK97085.1 ATLV01020986 KE525311 KFB45950.1 PNF38947.1 EAT40292.1 APCN01004968 CH477450 EJY57695.1 ABLF02031950 ABLF02031955 AXCM01003236 CRL05180.1 KQ434803 KZC05938.1
SOQ48987.1 AK402125 BAM18747.1 RSAL01000098 RVE47611.1 RVE47614.1 AGBW02010816 OWR47692.1 AGBW02013495 OWR43336.1 ODYU01011151 SOQ56716.1 RVE47596.1 NWSH01001694 PCG70405.1 KQ460473 KPJ14444.1 RVE47617.1 PCG70404.1 AGBW02014853 OWR40990.1 KPJ14443.1 AK402124 BAM18746.1 RVE47613.1 KQ459591 KPI97101.1 RSAL01000002 RVE54897.1 KQ459600 KPI94302.1 NWSH01000865 PCG73808.1 KZ150313 PZC71449.1 AK402502 BAM19124.1 ODYU01002797 SOQ40740.1 JN033724 AEW46877.1 RVE47618.1 AK403997 BAM20084.1 AGBW02010907 OWR47533.1 KQ459995 KPJ18524.1 KQ460736 KPJ12593.1 DS234988 EEB09873.1 ATLV01017365 KE525164 KFB42149.1 AF053921 AAD21841.1 CVRI01000064 CRL05174.1 KK852411 KDR24454.1 KQ458725 KPJ05344.1 CH477477 EAT40291.1 GEZM01092351 JAV56586.1 GAPW01002075 JAC11523.1 NEVH01005287 PNF38945.1 APCN01003246 GAPW01002076 JAC11522.1 AAAB01008888 EGK97085.1 ATLV01020986 KE525311 KFB45950.1 PNF38947.1 EAT40292.1 APCN01004968 CH477450 EJY57695.1 ABLF02031950 ABLF02031955 AXCM01003236 CRL05180.1 KQ434803 KZC05938.1
Proteomes
Pfam
Interpro
IPR009003
Peptidase_S1_PA
+ More
IPR001314 Peptidase_S1A
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR033116 TRYPSIN_SER
IPR006029 Neurotrans-gated_channel_TM
IPR006202 Neur_chan_lig-bd
IPR036734 Neur_chan_lig-bd_sf
IPR036719 Neuro-gated_channel_TM_sf
IPR018000 Neurotransmitter_ion_chnl_CS
IPR006201 Neur_channel
IPR022700 CLIP
IPR005304 Rbsml_bgen_MeTrfase_EMG1/NEP1
IPR029028 Alpha/beta_knot_MTases
IPR029026 tRNA_m1G_MTases_N
IPR034750 CULT
IPR015947 PUA-like_sf
IPR003111 Lon_substr-bd
IPR004910 Yippee/Mis18/Cereblon
IPR001314 Peptidase_S1A
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR033116 TRYPSIN_SER
IPR006029 Neurotrans-gated_channel_TM
IPR006202 Neur_chan_lig-bd
IPR036734 Neur_chan_lig-bd_sf
IPR036719 Neuro-gated_channel_TM_sf
IPR018000 Neurotransmitter_ion_chnl_CS
IPR006201 Neur_channel
IPR022700 CLIP
IPR005304 Rbsml_bgen_MeTrfase_EMG1/NEP1
IPR029028 Alpha/beta_knot_MTases
IPR029026 tRNA_m1G_MTases_N
IPR034750 CULT
IPR015947 PUA-like_sf
IPR003111 Lon_substr-bd
IPR004910 Yippee/Mis18/Cereblon
SUPFAM
Gene 3D
ProteinModelPortal
H9JSB6
Q1HPQ6
Q5MPC5
A0A2H1W7F8
I4DLF7
A0A3S2NHC8
+ More
A0A437BB50 A0A212F1T1 A0A212EPD4 A0A2H1WUS1 A0A3S2LZB7 A0A2A4JE90 A0A194RAB5 A0A437BB20 A0A2A4JFQ7 A0A212EHP9 A0A194R9C5 I4DLF6 A0A437BB81 A0A194PUL8 A0A437BWN7 A0A194PLU8 A0A2A4JQ23 A0A2W1B5H2 I4DMI4 A0A2H1VIS4 G9F9H9 A0A437BB41 I4DQ94 A0A212F1E7 A0A194RM65 A0A0N0PBX6 E0V917 A0A084VW05 Q9XY63 A0A1J1J158 A0A067RX29 A0A0N1PHM7 A0A1S4FI15 Q16ZZ9 A0A1Y1KAU3 A0A1Y9HED1 A0A182JGW6 A0A023EQX6 A0A2J7RDN5 A0A2C9GS29 A0A023EPR9 F5HLC4 A0A084W6V6 A0A2J7RDN0 Q170A0 A0A182ID15 A0A182VFE5 A0A182X8T2 J9HYX9 X1WIF2 A0A182LU74 A0A182U5J8 A0A1J1J052 A0A154P3Y1
A0A437BB50 A0A212F1T1 A0A212EPD4 A0A2H1WUS1 A0A3S2LZB7 A0A2A4JE90 A0A194RAB5 A0A437BB20 A0A2A4JFQ7 A0A212EHP9 A0A194R9C5 I4DLF6 A0A437BB81 A0A194PUL8 A0A437BWN7 A0A194PLU8 A0A2A4JQ23 A0A2W1B5H2 I4DMI4 A0A2H1VIS4 G9F9H9 A0A437BB41 I4DQ94 A0A212F1E7 A0A194RM65 A0A0N0PBX6 E0V917 A0A084VW05 Q9XY63 A0A1J1J158 A0A067RX29 A0A0N1PHM7 A0A1S4FI15 Q16ZZ9 A0A1Y1KAU3 A0A1Y9HED1 A0A182JGW6 A0A023EQX6 A0A2J7RDN5 A0A2C9GS29 A0A023EPR9 F5HLC4 A0A084W6V6 A0A2J7RDN0 Q170A0 A0A182ID15 A0A182VFE5 A0A182X8T2 J9HYX9 X1WIF2 A0A182LU74 A0A182U5J8 A0A1J1J052 A0A154P3Y1
PDB
3W94
E-value=4.60294e-31,
Score=336
Ontologies
GO
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.716077
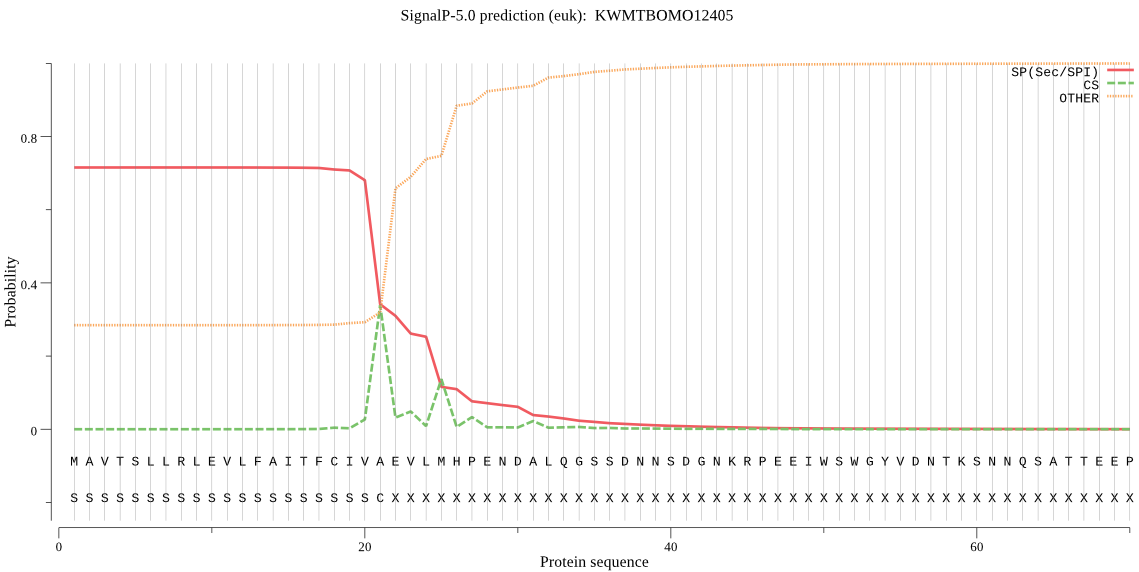
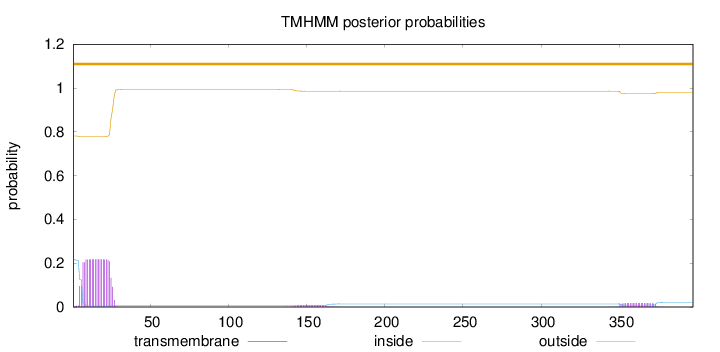
Length:
397
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.93716
Exp number, first 60 AAs:
4.33947
Total prob of N-in:
0.21952
outside
1 - 397
Population Genetic Test Statistics
Pi
265.902205
Theta
150.517313
Tajima's D
0.439725
CLR
1.194053
CSRT
0.503774811259437
Interpretation
Uncertain
