Gene
KWMTBOMO12402
Annotation
PREDICTED:_arrestin_domain-containing_protein_2-like_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.063 PlasmaMembrane Reliability : 1.648
Sequence
CDS
ATGACTTGGGATAATTGTATAGTACGATTAAACAGTGATTCTACGGGAAGTTTTTATACTGGTGACATAGTTACCGGAACCGTGATATTAGAATTTGACCAAGAACAAAAAATTGAAAGAATCGATTTACATGTTATAGGTATAAGCAAAGCACAATGGACCAGATCCATGCCCACTATGCCATATATTAAAATTTACTCAGAAAAGAAAAAAATATTAAGCATTAGCATTTGTGACATTTTTAGTGAAATAATTAGTGGTAAAAAGATTAGACCGGGTATCTACACTTACCCTTTCCACTTCGCTCTCCCCTTAGATTTACCATCTAGTTTCGAAAGCAAGATTGCTAAAGTGCACTATTGTATTAAAATTAAAAGCAAACCTGCGGTCAAAGTAAGAAAGAACGTTCCTCTGCGAATTCTGGAAAATGTCAACTTGAATCATTTGGAGGAAATGATGATAACATCGATCCACGACTTCGTAAAAGCATTTAGGACTTCAGGAAAATTCTCGGTTTCTGTGAAAACCTATAGGGCATTCGCATCCAAACAGACTGTCCCATTTGAGATCATATTAAACAATAGTAGAAAAGTAAAAATTAGTAAATTAACAGTTGCATTAATACAGAAACTAAGGTACGAAGTCCAGACAGGATATGCGGAGGAGGAGAAGACAATGTGTAAAGCGGAAAATAAGAAATTTGCGAATGCTTTAGTCGAGATCTGTAACTTAAAAATAGAAATGCCATCATTATGCCCTTCTACTATTCACTCGCTTGGTGCGATGGTTAACATTTCATATGAGTTCAGGGTAAAAGTAAAATTTCGATTTCATTTTTCCTTGATTGGAAGCATACCAGTGACAGTGGCAACAGTGCCTGTGATACACCATGATTTTTGA
Protein
MTWDNCIVRLNSDSTGSFYTGDIVTGTVILEFDQEQKIERIDLHVIGISKAQWTRSMPTMPYIKIYSEKKKILSISICDIFSEIISGKKIRPGIYTYPFHFALPLDLPSSFESKIAKVHYCIKIKSKPAVKVRKNVPLRILENVNLNHLEEMMITSIHDFVKAFRTSGKFSVSVKTYRAFASKQTVPFEIILNNSRKVKISKLTVALIQKLRYEVQTGYAEEEKTMCKAENKKFANALVEICNLKIEMPSLCPSTIHSLGAMVNISYEFRVKVKFRFHFSLIGSIPVTVATVPVIHHDF
Summary
Uniprot
A0A2A4JLV4
A0A2H1WAS3
A0A1E1W2P9
A0A1E1WJ31
A0A3S2NCL0
A0A194PHG2
+ More
A0A1E1WIW9 A0A194R0S7 A0A084VRX4 A0A1I8Q2Q7 A0A1I8MNN9 Q294M1 B4GMX0 A0A1B0FMP6 A0A1A9ZAA5 B3MTN5 A0A1B0BKC1 A0A1A9VW93 A0A0R3NJV2 A0A3B0JPD1 A0A1A9YAU1 A0A0P8XTR0 A0A0P8ZDS2 A0A1A9ZRT1 A0A1A9W1M2 A0A0K8VCF3 A0A3B0JSS8 A0A0K8UUR9 A0A1L8HN40 A0A182R0Y1 A0A034WVV7 A0A182KM46 A0A1B0C6S1 A0A1I8P3Z9 W8BY55 A0A1A9YAW6 A0A1W4VYB2 W8BHM6 A0A182TER2 Q7PYE1 A0A182IIG2 A0A182V8I5 B4I4X6 A0A1W4VLX5 B3NYR8 B4R099 B4IUW0 A0A182VQX7 A0A0R1E8K1 A0A0R1EE99 B4JHJ4 A0A0M5J0K9 Q16ZJ0 A0A240PMW7 A0A084VRX3 A0A182G1R7 B4M4K4 A0A023ERS7 B4N8B3 A0A1B0EUE3 Q9VHZ7 A0A0B4KGR0 Q6GR39 A0A0B4KGD9 B4I4W8 B4K6T5 A0A2M3Z758 A0A0Q9X012 A0A0Q9X4R9 A8JQV1 A0A182XBL9 A0A0M5J2I5 A0A1B0FFE3 A0A2M4BP22 B4N8C1 W5J2A4 A0A2M4AK63 A0A182N5X3 B3NYR1 A0A0A1WEU9 A0A1L8DN73 A0A0K8VTN5 A0A034VNU2 A0A182MAM7 A0A336MW53 A0A182R1I5 A0A087ZZ60 A0A084VRW8 A0A034VJJ4 W8BV05 B0WMD4 B4NAB2 A0A1J1IJ67 A0A0R1EFE1 Q08CX7 Q294M8 A0A1J1IGC8 A0A023ET25 B4JF67 A0A182PC22
A0A1E1WIW9 A0A194R0S7 A0A084VRX4 A0A1I8Q2Q7 A0A1I8MNN9 Q294M1 B4GMX0 A0A1B0FMP6 A0A1A9ZAA5 B3MTN5 A0A1B0BKC1 A0A1A9VW93 A0A0R3NJV2 A0A3B0JPD1 A0A1A9YAU1 A0A0P8XTR0 A0A0P8ZDS2 A0A1A9ZRT1 A0A1A9W1M2 A0A0K8VCF3 A0A3B0JSS8 A0A0K8UUR9 A0A1L8HN40 A0A182R0Y1 A0A034WVV7 A0A182KM46 A0A1B0C6S1 A0A1I8P3Z9 W8BY55 A0A1A9YAW6 A0A1W4VYB2 W8BHM6 A0A182TER2 Q7PYE1 A0A182IIG2 A0A182V8I5 B4I4X6 A0A1W4VLX5 B3NYR8 B4R099 B4IUW0 A0A182VQX7 A0A0R1E8K1 A0A0R1EE99 B4JHJ4 A0A0M5J0K9 Q16ZJ0 A0A240PMW7 A0A084VRX3 A0A182G1R7 B4M4K4 A0A023ERS7 B4N8B3 A0A1B0EUE3 Q9VHZ7 A0A0B4KGR0 Q6GR39 A0A0B4KGD9 B4I4W8 B4K6T5 A0A2M3Z758 A0A0Q9X012 A0A0Q9X4R9 A8JQV1 A0A182XBL9 A0A0M5J2I5 A0A1B0FFE3 A0A2M4BP22 B4N8C1 W5J2A4 A0A2M4AK63 A0A182N5X3 B3NYR1 A0A0A1WEU9 A0A1L8DN73 A0A0K8VTN5 A0A034VNU2 A0A182MAM7 A0A336MW53 A0A182R1I5 A0A087ZZ60 A0A084VRW8 A0A034VJJ4 W8BV05 B0WMD4 B4NAB2 A0A1J1IJ67 A0A0R1EFE1 Q08CX7 Q294M8 A0A1J1IGC8 A0A023ET25 B4JF67 A0A182PC22
Pubmed
EMBL
NWSH01001088
PCG72698.1
ODYU01007422
SOQ50179.1
GDQN01009808
JAT81246.1
+ More
GDQN01004062 JAT86992.1 RSAL01000098 RVE47623.1 KQ459603 KPI92851.1 GDQN01004091 JAT86963.1 KQ460883 KPJ11114.1 ATLV01015783 KE525036 KFB40718.1 CM000070 EAL28939.1 CH479185 EDW38194.1 CCAG010020238 CCAG010020239 CH902623 EDV30166.1 JXJN01015880 KRT01179.1 OUUW01000008 SPP84074.1 KPU72713.1 KPU72712.1 GDHF01015778 JAI36536.1 SPP84073.1 GDHF01021897 JAI30417.1 CM004467 OCT97512.1 AXCN02000190 GAKP01001024 JAC57928.1 JXJN01026881 GAMC01008314 JAB98241.1 GAMC01008313 JAB98242.1 AAAB01008987 EAA01049.4 APCN01002073 CH480821 EDW55269.1 CH954181 EDV48181.1 CM000364 EDX12024.1 CH892153 EDX00174.1 KRK05465.1 KRK05466.1 CH916369 EDV92821.1 CP012526 ALC47986.1 CH477489 EAT40084.1 KFB40717.1 JXUM01038907 JXUM01038908 JXUM01038909 KQ561184 KXJ79435.1 CH940652 EDW59565.2 JXUM01076838 JXUM01076839 GAPW01001678 KQ562972 JAC11920.1 KXJ74731.1 CH964232 EDW81364.1 AJWK01013619 AE014297 AY051771 AAF54149.3 AAK93195.1 AGB95730.1 BC071094 CM004466 AAH71094.1 OCU00426.1 AGB95731.1 EDW55261.1 CH933806 EDW14201.1 GGFM01003527 MBW24278.1 KRG00905.1 KRG00906.1 ABW08616.1 ALC47386.1 CCAG010003162 GGFJ01005695 MBW54836.1 EDW81372.1 ADMH02002134 ETN58387.1 GGFK01007852 MBW41173.1 EDV48174.1 GBXI01017354 JAC96937.1 GFDF01006213 JAV07871.1 GDHF01010384 JAI41930.1 GAKP01015477 GAKP01015474 GAKP01015472 GAKP01015467 GAKP01015464 JAC43485.1 AXCM01003758 UFQT01002599 SSX33751.1 KFB40712.1 GAKP01015476 JAC43476.1 GAMC01005752 GAMC01005751 JAC00804.1 DS231997 EDS30991.1 EDW81799.1 CVRI01000051 CRK99588.1 CH892151 KRK05463.1 AAMC01084980 BC124042 BC160577 AAI24043.1 AAI60577.1 EAL28938.1 CVRI01000048 CRK98596.1 GAPW01002079 JAC11519.1 EDV93348.1
GDQN01004062 JAT86992.1 RSAL01000098 RVE47623.1 KQ459603 KPI92851.1 GDQN01004091 JAT86963.1 KQ460883 KPJ11114.1 ATLV01015783 KE525036 KFB40718.1 CM000070 EAL28939.1 CH479185 EDW38194.1 CCAG010020238 CCAG010020239 CH902623 EDV30166.1 JXJN01015880 KRT01179.1 OUUW01000008 SPP84074.1 KPU72713.1 KPU72712.1 GDHF01015778 JAI36536.1 SPP84073.1 GDHF01021897 JAI30417.1 CM004467 OCT97512.1 AXCN02000190 GAKP01001024 JAC57928.1 JXJN01026881 GAMC01008314 JAB98241.1 GAMC01008313 JAB98242.1 AAAB01008987 EAA01049.4 APCN01002073 CH480821 EDW55269.1 CH954181 EDV48181.1 CM000364 EDX12024.1 CH892153 EDX00174.1 KRK05465.1 KRK05466.1 CH916369 EDV92821.1 CP012526 ALC47986.1 CH477489 EAT40084.1 KFB40717.1 JXUM01038907 JXUM01038908 JXUM01038909 KQ561184 KXJ79435.1 CH940652 EDW59565.2 JXUM01076838 JXUM01076839 GAPW01001678 KQ562972 JAC11920.1 KXJ74731.1 CH964232 EDW81364.1 AJWK01013619 AE014297 AY051771 AAF54149.3 AAK93195.1 AGB95730.1 BC071094 CM004466 AAH71094.1 OCU00426.1 AGB95731.1 EDW55261.1 CH933806 EDW14201.1 GGFM01003527 MBW24278.1 KRG00905.1 KRG00906.1 ABW08616.1 ALC47386.1 CCAG010003162 GGFJ01005695 MBW54836.1 EDW81372.1 ADMH02002134 ETN58387.1 GGFK01007852 MBW41173.1 EDV48174.1 GBXI01017354 JAC96937.1 GFDF01006213 JAV07871.1 GDHF01010384 JAI41930.1 GAKP01015477 GAKP01015474 GAKP01015472 GAKP01015467 GAKP01015464 JAC43485.1 AXCM01003758 UFQT01002599 SSX33751.1 KFB40712.1 GAKP01015476 JAC43476.1 GAMC01005752 GAMC01005751 JAC00804.1 DS231997 EDS30991.1 EDW81799.1 CVRI01000051 CRK99588.1 CH892151 KRK05463.1 AAMC01084980 BC124042 BC160577 AAI24043.1 AAI60577.1 EAL28938.1 CVRI01000048 CRK98596.1 GAPW01002079 JAC11519.1 EDV93348.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000030765
UP000095300
+ More
UP000095301 UP000001819 UP000008744 UP000092444 UP000092445 UP000007801 UP000092460 UP000078200 UP000268350 UP000092443 UP000091820 UP000186698 UP000075886 UP000075882 UP000192221 UP000075902 UP000007062 UP000075840 UP000075903 UP000001292 UP000008711 UP000000304 UP000002282 UP000075920 UP000001070 UP000092553 UP000008820 UP000075881 UP000069940 UP000249989 UP000008792 UP000007798 UP000092461 UP000000803 UP000009192 UP000076407 UP000000673 UP000075884 UP000075883 UP000075900 UP000005203 UP000002320 UP000183832 UP000008143 UP000075885
UP000095301 UP000001819 UP000008744 UP000092444 UP000092445 UP000007801 UP000092460 UP000078200 UP000268350 UP000092443 UP000091820 UP000186698 UP000075886 UP000075882 UP000192221 UP000075902 UP000007062 UP000075840 UP000075903 UP000001292 UP000008711 UP000000304 UP000002282 UP000075920 UP000001070 UP000092553 UP000008820 UP000075881 UP000069940 UP000249989 UP000008792 UP000007798 UP000092461 UP000000803 UP000009192 UP000076407 UP000000673 UP000075884 UP000075883 UP000075900 UP000005203 UP000002320 UP000183832 UP000008143 UP000075885
PRIDE
Interpro
SUPFAM
SSF81296
SSF81296
Gene 3D
ProteinModelPortal
A0A2A4JLV4
A0A2H1WAS3
A0A1E1W2P9
A0A1E1WJ31
A0A3S2NCL0
A0A194PHG2
+ More
A0A1E1WIW9 A0A194R0S7 A0A084VRX4 A0A1I8Q2Q7 A0A1I8MNN9 Q294M1 B4GMX0 A0A1B0FMP6 A0A1A9ZAA5 B3MTN5 A0A1B0BKC1 A0A1A9VW93 A0A0R3NJV2 A0A3B0JPD1 A0A1A9YAU1 A0A0P8XTR0 A0A0P8ZDS2 A0A1A9ZRT1 A0A1A9W1M2 A0A0K8VCF3 A0A3B0JSS8 A0A0K8UUR9 A0A1L8HN40 A0A182R0Y1 A0A034WVV7 A0A182KM46 A0A1B0C6S1 A0A1I8P3Z9 W8BY55 A0A1A9YAW6 A0A1W4VYB2 W8BHM6 A0A182TER2 Q7PYE1 A0A182IIG2 A0A182V8I5 B4I4X6 A0A1W4VLX5 B3NYR8 B4R099 B4IUW0 A0A182VQX7 A0A0R1E8K1 A0A0R1EE99 B4JHJ4 A0A0M5J0K9 Q16ZJ0 A0A240PMW7 A0A084VRX3 A0A182G1R7 B4M4K4 A0A023ERS7 B4N8B3 A0A1B0EUE3 Q9VHZ7 A0A0B4KGR0 Q6GR39 A0A0B4KGD9 B4I4W8 B4K6T5 A0A2M3Z758 A0A0Q9X012 A0A0Q9X4R9 A8JQV1 A0A182XBL9 A0A0M5J2I5 A0A1B0FFE3 A0A2M4BP22 B4N8C1 W5J2A4 A0A2M4AK63 A0A182N5X3 B3NYR1 A0A0A1WEU9 A0A1L8DN73 A0A0K8VTN5 A0A034VNU2 A0A182MAM7 A0A336MW53 A0A182R1I5 A0A087ZZ60 A0A084VRW8 A0A034VJJ4 W8BV05 B0WMD4 B4NAB2 A0A1J1IJ67 A0A0R1EFE1 Q08CX7 Q294M8 A0A1J1IGC8 A0A023ET25 B4JF67 A0A182PC22
A0A1E1WIW9 A0A194R0S7 A0A084VRX4 A0A1I8Q2Q7 A0A1I8MNN9 Q294M1 B4GMX0 A0A1B0FMP6 A0A1A9ZAA5 B3MTN5 A0A1B0BKC1 A0A1A9VW93 A0A0R3NJV2 A0A3B0JPD1 A0A1A9YAU1 A0A0P8XTR0 A0A0P8ZDS2 A0A1A9ZRT1 A0A1A9W1M2 A0A0K8VCF3 A0A3B0JSS8 A0A0K8UUR9 A0A1L8HN40 A0A182R0Y1 A0A034WVV7 A0A182KM46 A0A1B0C6S1 A0A1I8P3Z9 W8BY55 A0A1A9YAW6 A0A1W4VYB2 W8BHM6 A0A182TER2 Q7PYE1 A0A182IIG2 A0A182V8I5 B4I4X6 A0A1W4VLX5 B3NYR8 B4R099 B4IUW0 A0A182VQX7 A0A0R1E8K1 A0A0R1EE99 B4JHJ4 A0A0M5J0K9 Q16ZJ0 A0A240PMW7 A0A084VRX3 A0A182G1R7 B4M4K4 A0A023ERS7 B4N8B3 A0A1B0EUE3 Q9VHZ7 A0A0B4KGR0 Q6GR39 A0A0B4KGD9 B4I4W8 B4K6T5 A0A2M3Z758 A0A0Q9X012 A0A0Q9X4R9 A8JQV1 A0A182XBL9 A0A0M5J2I5 A0A1B0FFE3 A0A2M4BP22 B4N8C1 W5J2A4 A0A2M4AK63 A0A182N5X3 B3NYR1 A0A0A1WEU9 A0A1L8DN73 A0A0K8VTN5 A0A034VNU2 A0A182MAM7 A0A336MW53 A0A182R1I5 A0A087ZZ60 A0A084VRW8 A0A034VJJ4 W8BV05 B0WMD4 B4NAB2 A0A1J1IJ67 A0A0R1EFE1 Q08CX7 Q294M8 A0A1J1IGC8 A0A023ET25 B4JF67 A0A182PC22
PDB
4R7X
E-value=3.29086e-05,
Score=111
Ontologies
GO
Topology
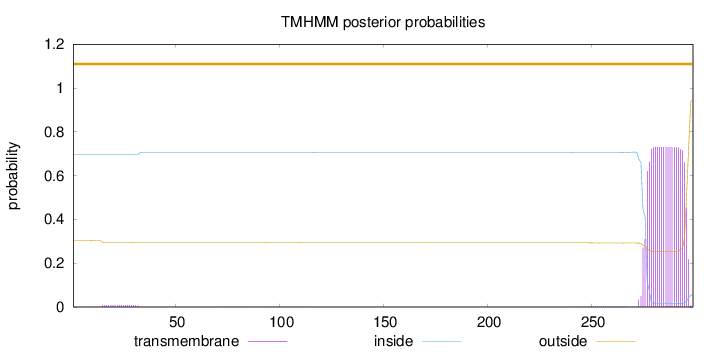
Length:
299
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
15.16827
Exp number, first 60 AAs:
0.15824
Total prob of N-in:
0.69681
outside
1 - 299
Population Genetic Test Statistics
Pi
203.191672
Theta
171.07913
Tajima's D
0.445949
CLR
0.702404
CSRT
0.491375431228439
Interpretation
Uncertain
