Gene
KWMTBOMO12398
Pre Gene Modal
BGIBMGA012442
Annotation
PREDICTED:_ankyrin_repeat_domain-containing_protein_29-like_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.512 Extracellular Reliability : 1.334
Sequence
CDS
ATGTACGGGAGGGCGCCAATACACTGGGCCGCCAGCCGTGGGAACGTTGAAATAATCAACTTACTTATCGAAGCCAAGTGCGATATAGAAGCTGCTGATAAGTTCGGGATGCGGCCGTTGCTGATGGCGGCTTGGCATGGCCATCTAGACGCCGTGACATCTCTAGTCAAGGCAGGCGCATGTCTCGCAGCTACTAACAAAAAGCATAATGATCTCCTGCAATGTGCGTGTGCCCGCGGCGCCCGCGATGTGGCGGAATACGCCCTGTCCCTCGGAGCGCGCGTGCAGACTGCAGACGTCGCTGGAGACGCCCCCCTCGCACAAGCTGCAAGAGCTGGACACGCGGCTGTCGTAGAACTACTCCTGGACAAGGGGGCTGATATCGATGCAGTCAATAAGAACGGTCGTACGGCTCTACACATGGCTTGCGAAGGCGGATACTCCTCAACTGCTAGTCTCCTGGTATCACGAGGTGCGTCTAAGGAAGCCAGGGACAACGCCGGCAGGACACCAATACACATAGCTGCAGTCCACAGACACACGGAGCTCGTGAGGGTCCTTTTGGAGGCGGAATGCTTCGTCGATGCGGTTGATGATAGCGGTGTGACCGCGCTACAACTGGCGTGTGCACAAGGCTGCCGGGGCATCGCGGAGCACCTGCTTGAGTTTGGGGCAAATGTTCACTTACAGAATAACGTAGGTTCCTCGGCTTTGCACGCCGCGTGCGCCGCCGACGCAAATGACATTGTGGAGCTGCTATTAGCCAACGGAGCAGATCCCAGCCTCACCGACCAGCCGTCGAAGTTAACCGCATTCATAATGCTAGGCAGCAGCTTGGCTCTGCGCCTGGCATTGCTGAAGTCCATGGGCGACGGTAATCACTCACCATCAGGTGGGCCGTATGCTCGTCTGCATACAAGGGCAATAAAATAA
Protein
MYGRAPIHWAASRGNVEIINLLIEAKCDIEAADKFGMRPLLMAAWHGHLDAVTSLVKAGACLAATNKKHNDLLQCACARGARDVAEYALSLGARVQTADVAGDAPLAQAARAGHAAVVELLLDKGADIDAVNKNGRTALHMACEGGYSSTASLLVSRGASKEARDNAGRTPIHIAAVHRHTELVRVLLEAECFVDAVDDSGVTALQLACAQGCRGIAEHLLEFGANVHLQNNVGSSALHAACAADANDIVELLLANGADPSLTDQPSKLTAFIMLGSSLALRLALLKSMGDGNHSPSGGPYARLHTRAIK
Summary
Similarity
Belongs to the peptidase S8 family.
Uniprot
A0A194R9Z2
A0A194Q120
A0A437BB80
A0A2A4JMR1
A0A2A4JMR6
A0A2A4JL46
+ More
A0A0L7L4V0 A0A1Y1M2D4 D6X569 A0A2J7Q334 A0A1B6KFU8 A0A1B6MGG7 A0A1B6CEW9 A0A2S2P6T4 A0A2H8TID9 J9K952 A0A0T6B8J8 A0A3L8DLG0 A0A026W7K6 E2B514 A0A087ZN61 A0A151XG86 F4X0J3 A0A158NDE3 A0A0L7R6U4 A0A154PEX7 A0A195B8T5 A0A195DV78 A0A151IDR1 A0A195F7P7 E2AV01 T1E1B2 A0A094HCR5 A0A355KTI7 A0A1V5PQL8 T1KXU1 A0A3D8Q7F7 T1JDD6 G9NK45 B7PMP3 A0A2H3TWG0 A0A094B9X5 A0A429GKC2 A0A094I596 A2DPI2 A2H1V6 X0H183 A0A1V9Z2B8 U1HXR6 D3B1S8 A0A1S3HS58 W4YMA5 A2FTQ7 B3EU24 A0A3L6NP46 A0A1X6P331 W4XCZ4 A0A420PX84 A0A452HAR4 X0BB66 A0A219ARV3 X0C681 A0A3D8SHD6 X0KFW7 A0A1Q9DQ07 A0A1L7WB49 A2GUQ1 A0A3D8S806 A2DJV1 A0A094GWC4 A2F776 J9HSB7 A0A1S7UK06 W4YMA4 A0A2H3GF88 A0A178Z6K5 A0A094GHK3 A2EDV4 A0A420PI16 A0A0D9NI83 A0A150FY68 B8NAF0 A0A3L5TPW1 A0A369RGB3 B8MPS3 A0A351KWZ6 A0A2G7FJA5 A0A3M7JWA9 A0A2E3MI11 A0A2P1P8I2 L8G7R4 A0A1S9DC68 A0A1L7UFU6 A0A2J7ZNP1 K1Q8R1
A0A0L7L4V0 A0A1Y1M2D4 D6X569 A0A2J7Q334 A0A1B6KFU8 A0A1B6MGG7 A0A1B6CEW9 A0A2S2P6T4 A0A2H8TID9 J9K952 A0A0T6B8J8 A0A3L8DLG0 A0A026W7K6 E2B514 A0A087ZN61 A0A151XG86 F4X0J3 A0A158NDE3 A0A0L7R6U4 A0A154PEX7 A0A195B8T5 A0A195DV78 A0A151IDR1 A0A195F7P7 E2AV01 T1E1B2 A0A094HCR5 A0A355KTI7 A0A1V5PQL8 T1KXU1 A0A3D8Q7F7 T1JDD6 G9NK45 B7PMP3 A0A2H3TWG0 A0A094B9X5 A0A429GKC2 A0A094I596 A2DPI2 A2H1V6 X0H183 A0A1V9Z2B8 U1HXR6 D3B1S8 A0A1S3HS58 W4YMA5 A2FTQ7 B3EU24 A0A3L6NP46 A0A1X6P331 W4XCZ4 A0A420PX84 A0A452HAR4 X0BB66 A0A219ARV3 X0C681 A0A3D8SHD6 X0KFW7 A0A1Q9DQ07 A0A1L7WB49 A2GUQ1 A0A3D8S806 A2DJV1 A0A094GWC4 A2F776 J9HSB7 A0A1S7UK06 W4YMA4 A0A2H3GF88 A0A178Z6K5 A0A094GHK3 A2EDV4 A0A420PI16 A0A0D9NI83 A0A150FY68 B8NAF0 A0A3L5TPW1 A0A369RGB3 B8MPS3 A0A351KWZ6 A0A2G7FJA5 A0A3M7JWA9 A0A2E3MI11 A0A2P1P8I2 L8G7R4 A0A1S9DC68 A0A1L7UFU6 A0A2J7ZNP1 K1Q8R1
Pubmed
EMBL
KQ460473
KPJ14452.1
KQ459591
KPI97095.1
RSAL01000098
RVE47625.1
+ More
NWSH01001088 PCG72692.1 PCG72690.1 PCG72691.1 JTDY01002914 KOB70497.1 GEZM01042394 JAV79889.1 KQ971382 EEZ97168.2 NEVH01019070 PNF23005.1 GEBQ01029678 JAT10299.1 GEBQ01004982 JAT34995.1 GEDC01025316 GEDC01023929 JAS11982.1 JAS13369.1 GGMR01012453 MBY25072.1 GFXV01002069 MBW13874.1 ABLF02021976 ABLF02021979 ABLF02021982 ABLF02021985 LJIG01009204 KRT83547.1 QOIP01000007 RLU21043.1 KK107364 EZA51978.1 GL445685 EFN89222.1 KQ982173 KYQ59412.1 GL888498 EGI59985.1 ADTU01012193 ADTU01012194 ADTU01012195 ADTU01012196 KQ414646 KOC66568.1 KQ434890 KZC10409.1 KQ976558 KYM80647.1 KQ980341 KYN16469.1 KQ977906 KYM98863.1 KQ981744 KYN36406.1 GL442974 EFN62719.1 GAKT01000054 JAA93008.1 JPKE01002539 KFZ12697.1 DOAP01000002 HBL98858.1 MWAT01000023 OQA19887.1 CAEY01000697 PDLN01000022 RDW57753.1 JH432104 ABDG02000017 EHK49265.1 ABJB010077134 ABJB010289115 ABJB010395643 ABJB010819276 ABJB010872884 ABJB010968974 DS748694 EEC07865.1 FMJY01000007 SCO87656.1 JPJV01000174 KFY32155.1 RCOS01000097 RSN74274.1 JPKA01001201 KFY91707.1 DS113227 EAY17726.1 DS124314 EAX76611.1 JH659097 EXL65836.1 JNBS01002361 OQR92047.1 KE720882 ERF74254.1 ADBJ01000008 EFA85252.1 AAGJ04156956 DS114016 EAX91696.1 CP001102 ACE06726.1 MRCU01000004 RKK19744.1 KV918910 OSX75264.1 AAGJ04165197 MRCY01000114 RKK97114.1 JH658472 EXK79410.1 LSBJ02000002 OWT43362.1 JH658534 EXK78232.1 PDLM01000002 RDW85168.1 KK035598 EXM12393.1 LSRX01000441 OLP97247.1 FJOF01000018 CZR49696.1 DS120513 EAX79116.1 PDLM01000003 RDW82340.1 DS113209 EAY19315.1 JPKC01000293 KFZ00411.1 DS113644 EAX99243.1 CH477275 EJY57493.1 DF977447 GAP83599.2 MABQ02000011 PCD23152.1 LVYI01000011 OAP55332.1 JPKC01000049 KFZ02422.1 DS113363 EAY09170.1 MRCY01000202 RKK92160.1 KE384850 KJK73423.1 LSYV01000135 KXZ42561.1 EQ963476 EED52293.1 KV593670 OPL21239.1 QPIP01000119 RDD34098.1 EQ962659 EED12708.1 DNGJ01000009 HAZ43066.1 NEXV01000614 PIG80385.1 QQZZ01000110 RMZ41819.1 PAPY01000021 MBB71744.1 CP027845 AVP87590.1 GL573230 ELR09112.1 MKZY01000007 OOO06691.1 FCQH01000036 CVL09269.1 PGGS01000769 PNH01893.1 JH818280 EKC25260.1
NWSH01001088 PCG72692.1 PCG72690.1 PCG72691.1 JTDY01002914 KOB70497.1 GEZM01042394 JAV79889.1 KQ971382 EEZ97168.2 NEVH01019070 PNF23005.1 GEBQ01029678 JAT10299.1 GEBQ01004982 JAT34995.1 GEDC01025316 GEDC01023929 JAS11982.1 JAS13369.1 GGMR01012453 MBY25072.1 GFXV01002069 MBW13874.1 ABLF02021976 ABLF02021979 ABLF02021982 ABLF02021985 LJIG01009204 KRT83547.1 QOIP01000007 RLU21043.1 KK107364 EZA51978.1 GL445685 EFN89222.1 KQ982173 KYQ59412.1 GL888498 EGI59985.1 ADTU01012193 ADTU01012194 ADTU01012195 ADTU01012196 KQ414646 KOC66568.1 KQ434890 KZC10409.1 KQ976558 KYM80647.1 KQ980341 KYN16469.1 KQ977906 KYM98863.1 KQ981744 KYN36406.1 GL442974 EFN62719.1 GAKT01000054 JAA93008.1 JPKE01002539 KFZ12697.1 DOAP01000002 HBL98858.1 MWAT01000023 OQA19887.1 CAEY01000697 PDLN01000022 RDW57753.1 JH432104 ABDG02000017 EHK49265.1 ABJB010077134 ABJB010289115 ABJB010395643 ABJB010819276 ABJB010872884 ABJB010968974 DS748694 EEC07865.1 FMJY01000007 SCO87656.1 JPJV01000174 KFY32155.1 RCOS01000097 RSN74274.1 JPKA01001201 KFY91707.1 DS113227 EAY17726.1 DS124314 EAX76611.1 JH659097 EXL65836.1 JNBS01002361 OQR92047.1 KE720882 ERF74254.1 ADBJ01000008 EFA85252.1 AAGJ04156956 DS114016 EAX91696.1 CP001102 ACE06726.1 MRCU01000004 RKK19744.1 KV918910 OSX75264.1 AAGJ04165197 MRCY01000114 RKK97114.1 JH658472 EXK79410.1 LSBJ02000002 OWT43362.1 JH658534 EXK78232.1 PDLM01000002 RDW85168.1 KK035598 EXM12393.1 LSRX01000441 OLP97247.1 FJOF01000018 CZR49696.1 DS120513 EAX79116.1 PDLM01000003 RDW82340.1 DS113209 EAY19315.1 JPKC01000293 KFZ00411.1 DS113644 EAX99243.1 CH477275 EJY57493.1 DF977447 GAP83599.2 MABQ02000011 PCD23152.1 LVYI01000011 OAP55332.1 JPKC01000049 KFZ02422.1 DS113363 EAY09170.1 MRCY01000202 RKK92160.1 KE384850 KJK73423.1 LSYV01000135 KXZ42561.1 EQ963476 EED52293.1 KV593670 OPL21239.1 QPIP01000119 RDD34098.1 EQ962659 EED12708.1 DNGJ01000009 HAZ43066.1 NEXV01000614 PIG80385.1 QQZZ01000110 RMZ41819.1 PAPY01000021 MBB71744.1 CP027845 AVP87590.1 GL573230 ELR09112.1 MKZY01000007 OOO06691.1 FCQH01000036 CVL09269.1 PGGS01000769 PNH01893.1 JH818280 EKC25260.1
Proteomes
UP000053240
UP000053268
UP000283053
UP000218220
UP000037510
UP000007266
+ More
UP000235965 UP000007819 UP000279307 UP000053097 UP000008237 UP000005203 UP000075809 UP000007755 UP000005205 UP000053825 UP000076502 UP000078540 UP000078492 UP000078542 UP000078541 UP000000311 UP000029308 UP000015104 UP000256328 UP000005426 UP000001555 UP000219369 UP000029327 UP000277582 UP000029270 UP000001542 UP000030676 UP000243217 UP000001396 UP000085678 UP000007110 UP000001227 UP000270866 UP000285860 UP000291020 UP000030663 UP000078397 UP000256645 UP000030701 UP000183971 UP000029284 UP000008820 UP000054516 UP000219602 UP000078343 UP000054544 UP000075714 UP000001875 UP000253942 UP000001745 UP000262455 UP000231358 UP000275480 UP000224825 UP000011064 UP000190312 UP000184255 UP000005408
UP000235965 UP000007819 UP000279307 UP000053097 UP000008237 UP000005203 UP000075809 UP000007755 UP000005205 UP000053825 UP000076502 UP000078540 UP000078492 UP000078542 UP000078541 UP000000311 UP000029308 UP000015104 UP000256328 UP000005426 UP000001555 UP000219369 UP000029327 UP000277582 UP000029270 UP000001542 UP000030676 UP000243217 UP000001396 UP000085678 UP000007110 UP000001227 UP000270866 UP000285860 UP000291020 UP000030663 UP000078397 UP000256645 UP000030701 UP000183971 UP000029284 UP000008820 UP000054516 UP000219602 UP000078343 UP000054544 UP000075714 UP000001875 UP000253942 UP000001745 UP000262455 UP000231358 UP000275480 UP000224825 UP000011064 UP000190312 UP000184255 UP000005408
PRIDE
Pfam
PF12796 Ank_2
+ More
PF13606 Ank_3
PF00531 Death
PF00023 Ank
PF06985 HET
PF16470 S8_pro-domain
PF01483 P_proprotein
PF00082 Peptidase_S8
PF05729 NACHT
PF17106 NACHT_sigma
PF17107 SesA
PF00307 CH
PF16095 COR
PF04564 U-box
PF14479 HeLo
PF12697 Abhydrolase_6
PF01048 PNP_UDP_1
PF00069 Pkinase
PF12937 F-box-like
PF13606 Ank_3
PF00531 Death
PF00023 Ank
PF06985 HET
PF16470 S8_pro-domain
PF01483 P_proprotein
PF00082 Peptidase_S8
PF05729 NACHT
PF17106 NACHT_sigma
PF17107 SesA
PF00307 CH
PF16095 COR
PF04564 U-box
PF14479 HeLo
PF12697 Abhydrolase_6
PF01048 PNP_UDP_1
PF00069 Pkinase
PF12937 F-box-like
Interpro
IPR020683
Ankyrin_rpt-contain_dom
+ More
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR000488 Death_domain
IPR039194 AKD1A
IPR011029 DEATH-like_dom_sf
IPR010730 HET
IPR032815 S8_pro-domain
IPR034182 Kexin/furin
IPR023828 Peptidase_S8_Ser-AS
IPR008979 Galactose-bd-like_sf
IPR036852 Peptidase_S8/S53_dom_sf
IPR015500 Peptidase_S8_subtilisin-rel
IPR038466 S8_pro-domain_sf
IPR022398 Peptidase_S8_His-AS
IPR000209 Peptidase_S8/S53_dom
IPR023827 Peptidase_S8_Asp-AS
IPR002884 P_dom
IPR031353 NACHT_sigma
IPR027417 P-loop_NTPase
IPR007111 NACHT_NTPase
IPR031352 SesA
IPR001715 CH-domain
IPR036872 CH_dom_sf
IPR003096 SM22_calponin
IPR036388 WH-like_DNA-bd_sf
IPR036514 SGNH_hydro_sf
IPR032171 COR
IPR013083 Znf_RING/FYVE/PHD
IPR003613 Ubox_domain
IPR038305 HeLo_sf
IPR029498 HeLo_dom
IPR000073 AB_hydrolase_1
IPR029058 AB_hydrolase
IPR029071 Ubiquitin-like_domsf
IPR035994 Nucleoside_phosphorylase_sf
IPR000845 Nucleoside_phosphorylase_d
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR017441 Protein_kinase_ATP_BS
IPR001810 F-box_dom
IPR036047 F-box-like_dom_sf
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR000488 Death_domain
IPR039194 AKD1A
IPR011029 DEATH-like_dom_sf
IPR010730 HET
IPR032815 S8_pro-domain
IPR034182 Kexin/furin
IPR023828 Peptidase_S8_Ser-AS
IPR008979 Galactose-bd-like_sf
IPR036852 Peptidase_S8/S53_dom_sf
IPR015500 Peptidase_S8_subtilisin-rel
IPR038466 S8_pro-domain_sf
IPR022398 Peptidase_S8_His-AS
IPR000209 Peptidase_S8/S53_dom
IPR023827 Peptidase_S8_Asp-AS
IPR002884 P_dom
IPR031353 NACHT_sigma
IPR027417 P-loop_NTPase
IPR007111 NACHT_NTPase
IPR031352 SesA
IPR001715 CH-domain
IPR036872 CH_dom_sf
IPR003096 SM22_calponin
IPR036388 WH-like_DNA-bd_sf
IPR036514 SGNH_hydro_sf
IPR032171 COR
IPR013083 Znf_RING/FYVE/PHD
IPR003613 Ubox_domain
IPR038305 HeLo_sf
IPR029498 HeLo_dom
IPR000073 AB_hydrolase_1
IPR029058 AB_hydrolase
IPR029071 Ubiquitin-like_domsf
IPR035994 Nucleoside_phosphorylase_sf
IPR000845 Nucleoside_phosphorylase_d
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR017441 Protein_kinase_ATP_BS
IPR001810 F-box_dom
IPR036047 F-box-like_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A194R9Z2
A0A194Q120
A0A437BB80
A0A2A4JMR1
A0A2A4JMR6
A0A2A4JL46
+ More
A0A0L7L4V0 A0A1Y1M2D4 D6X569 A0A2J7Q334 A0A1B6KFU8 A0A1B6MGG7 A0A1B6CEW9 A0A2S2P6T4 A0A2H8TID9 J9K952 A0A0T6B8J8 A0A3L8DLG0 A0A026W7K6 E2B514 A0A087ZN61 A0A151XG86 F4X0J3 A0A158NDE3 A0A0L7R6U4 A0A154PEX7 A0A195B8T5 A0A195DV78 A0A151IDR1 A0A195F7P7 E2AV01 T1E1B2 A0A094HCR5 A0A355KTI7 A0A1V5PQL8 T1KXU1 A0A3D8Q7F7 T1JDD6 G9NK45 B7PMP3 A0A2H3TWG0 A0A094B9X5 A0A429GKC2 A0A094I596 A2DPI2 A2H1V6 X0H183 A0A1V9Z2B8 U1HXR6 D3B1S8 A0A1S3HS58 W4YMA5 A2FTQ7 B3EU24 A0A3L6NP46 A0A1X6P331 W4XCZ4 A0A420PX84 A0A452HAR4 X0BB66 A0A219ARV3 X0C681 A0A3D8SHD6 X0KFW7 A0A1Q9DQ07 A0A1L7WB49 A2GUQ1 A0A3D8S806 A2DJV1 A0A094GWC4 A2F776 J9HSB7 A0A1S7UK06 W4YMA4 A0A2H3GF88 A0A178Z6K5 A0A094GHK3 A2EDV4 A0A420PI16 A0A0D9NI83 A0A150FY68 B8NAF0 A0A3L5TPW1 A0A369RGB3 B8MPS3 A0A351KWZ6 A0A2G7FJA5 A0A3M7JWA9 A0A2E3MI11 A0A2P1P8I2 L8G7R4 A0A1S9DC68 A0A1L7UFU6 A0A2J7ZNP1 K1Q8R1
A0A0L7L4V0 A0A1Y1M2D4 D6X569 A0A2J7Q334 A0A1B6KFU8 A0A1B6MGG7 A0A1B6CEW9 A0A2S2P6T4 A0A2H8TID9 J9K952 A0A0T6B8J8 A0A3L8DLG0 A0A026W7K6 E2B514 A0A087ZN61 A0A151XG86 F4X0J3 A0A158NDE3 A0A0L7R6U4 A0A154PEX7 A0A195B8T5 A0A195DV78 A0A151IDR1 A0A195F7P7 E2AV01 T1E1B2 A0A094HCR5 A0A355KTI7 A0A1V5PQL8 T1KXU1 A0A3D8Q7F7 T1JDD6 G9NK45 B7PMP3 A0A2H3TWG0 A0A094B9X5 A0A429GKC2 A0A094I596 A2DPI2 A2H1V6 X0H183 A0A1V9Z2B8 U1HXR6 D3B1S8 A0A1S3HS58 W4YMA5 A2FTQ7 B3EU24 A0A3L6NP46 A0A1X6P331 W4XCZ4 A0A420PX84 A0A452HAR4 X0BB66 A0A219ARV3 X0C681 A0A3D8SHD6 X0KFW7 A0A1Q9DQ07 A0A1L7WB49 A2GUQ1 A0A3D8S806 A2DJV1 A0A094GWC4 A2F776 J9HSB7 A0A1S7UK06 W4YMA4 A0A2H3GF88 A0A178Z6K5 A0A094GHK3 A2EDV4 A0A420PI16 A0A0D9NI83 A0A150FY68 B8NAF0 A0A3L5TPW1 A0A369RGB3 B8MPS3 A0A351KWZ6 A0A2G7FJA5 A0A3M7JWA9 A0A2E3MI11 A0A2P1P8I2 L8G7R4 A0A1S9DC68 A0A1L7UFU6 A0A2J7ZNP1 K1Q8R1
PDB
6MOL
E-value=3.34184e-30,
Score=327
Ontologies
GO
PANTHER
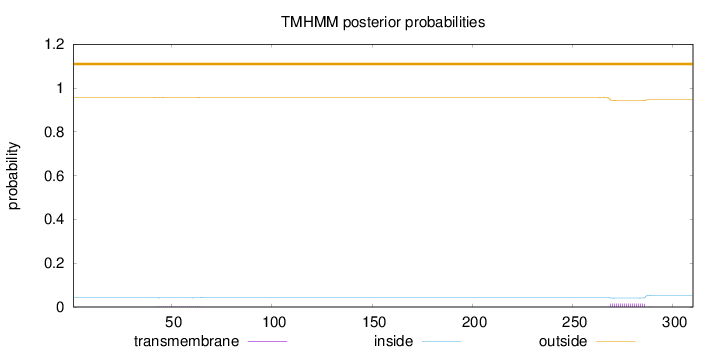
Topology
Length:
310
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.33139
Exp number, first 60 AAs:
0.0376
Total prob of N-in:
0.04204
outside
1 - 310
Population Genetic Test Statistics
Pi
26.611551
Theta
187.014591
Tajima's D
0.283018
CLR
0.073029
CSRT
0.449877506124694
Interpretation
Possibly Positive selection
