Gene
KWMTBOMO12394
Pre Gene Modal
BGIBMGA012432
Annotation
PREDICTED:_clavesin-2-like_isoform_X1_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.307 PlasmaMembrane Reliability : 1.85
Sequence
CDS
ATGCCGTTCATTGACATCGCCTTCCAAGCCGAACTAAGCCGATACGAAGACCCGGAGTTTGAAGAGTACGCCAAGAAGAATTGTAACGAATTCCCTGACACTAGGCATAAAGTAATCGAGGAGCTACGCCGCATGGTCCGCGAACGAGGTGAATGTATCCCTCGGCGCGTTGACGACGCATACTGTCTCAGATTTCTTCGATGTCGGCGGTTTATTCCTGCGTTAGCTCACAAATTGATGGTGCGTTACGAAGACTTTCGCCAAAAAAACGCGTACCTGTACAACTGTGACCCGTTTGGACTACAACGAGTTAAAAACGTCTACGGAGGTACTTTACCCGAAAGTCCGGAAAATGGAAGAATAGTTCTTATGAGGTTCGGACGCTGGGATCCCAGTGCTGTGCCGATCGAAGACGTGGTTCGTTGCGCCCTGTTCATGGATGAAATTGCTGTGATGCAGCCAAAACTACAGATCCTGGGCGTGACCATCATCGTAGACCTCGATGGGTTGAGCATCAGACACGTGAGGCATTTGACTCCGACAGTTGCCTACCAGATTGTCAGTCTTATGGGAGTTTCATTCCCGTTGGCGATGCACGGCCTCCACATCATAAACTACAACTGGATCTTGAATTCTGTGTTCTATTTGTTTAAACAATTTATACCGGCTGCAGTATGGAGCCGCCTGCATTTTCACGGCTCGAACATGGAATCACTCCACAAGTTCATACAGCCCCAGTACTTGCCACCCGAGTATGGGGGCAGTTGCAGACACATCATCTCGACTGAAGAATGGATCGCTAAGATCTATAGATACAAAGATGATTTCCTGATAGAAGAATTAAGAGATTTAGGTTTTTACGTTACATAG
Protein
MPFIDIAFQAELSRYEDPEFEEYAKKNCNEFPDTRHKVIEELRRMVRERGECIPRRVDDAYCLRFLRCRRFIPALAHKLMVRYEDFRQKNAYLYNCDPFGLQRVKNVYGGTLPESPENGRIVLMRFGRWDPSAVPIEDVVRCALFMDEIAVMQPKLQILGVTIIVDLDGLSIRHVRHLTPTVAYQIVSLMGVSFPLAMHGLHIINYNWILNSVFYLFKQFIPAAVWSRLHFHGSNMESLHKFIQPQYLPPEYGGSCRHIISTEEWIAKIYRYKDDFLIEELRDLGFYVT
Summary
Uniprot
H9JSC1
A0A194RAC5
A0A194PVS2
A0A2H4RMP6
A0A2W1BVU1
A0A1E1VYH6
+ More
A0A212F3L1 A0A2H1VEG3 A0A2A4JBT7 A0A1E1WU69 A0A437B2L8 A0A2A4J0W6 I4DLF8 A0A194PWG0 A0A2H4RMQ8 A0A194R9D4 A0A2H1V8Z2 A0A0S2Z3F3 H9JSD0 A0A212FK12 A0A0L7LCR3 A0A2H4RMU7 A0A2A4JCS5 A0A3S2NDL1 A0A0S2Z371 A0A2H4RMQ9 H9JSC0 A0A212EYW0 A0A2W1BQN2 A0A3S2LZG7 H9J0U5 S4PDY3 A0A2H4RMR4 A0A2H4RMU9 D6WK77 A0A0S2Z2X8 K7J584 A0A2J7PEI5 A0A437BEE0 A0A2A4J639 A0A1Q3FJS7 A0A151XFF6 A0A2C9H8Q1 A0A182U9P9 A0A182USL8 B0WIP9 E2C3S6 A0A453YJS9 A0A1S4H0R7 A0A1S4JIR2 A0A158NHN1 A0A212FFX7 A0A182QDV4 A0A2P8Y1N2 E9J2R5 A0A195D4M7 A0A182KCU4 A0A084VM70 A0A2M4BVL7 A0A182M7C8 A0A195AZZ6 A0A2H1WVH3 A0A2M4BVI7 A0A182SLW3 A0A1B6CWG4 A0A088A1P4 U5EUI3 F4WD80 A0A195EF31 A0A0M8ZXJ1 T1DG07 A0A182L6I3 A0A088AQZ8 A0A182JEP7 A0A2M3ZAW7 D6WK78 A0A0L7QVN0 E2B175 A0A195FWL3 A0A1Y1KBD3 W5JFN1 A0A1Y1KBD8 A0A2M4ADQ0 A0A2M4ADP7 A0A182YNG9 A0A194PPJ0 A0A1B6EH81 A0A1Y1MAD0 A0A0C9RS57 A0A1Y1M6Q3 D6WK79 A0A154PSY4 A0A069DR99 A0A182P9H0 E0VDE6 A0A1L8DYZ0 A0A026WVT7 A0A182VUK4
A0A212F3L1 A0A2H1VEG3 A0A2A4JBT7 A0A1E1WU69 A0A437B2L8 A0A2A4J0W6 I4DLF8 A0A194PWG0 A0A2H4RMQ8 A0A194R9D4 A0A2H1V8Z2 A0A0S2Z3F3 H9JSD0 A0A212FK12 A0A0L7LCR3 A0A2H4RMU7 A0A2A4JCS5 A0A3S2NDL1 A0A0S2Z371 A0A2H4RMQ9 H9JSC0 A0A212EYW0 A0A2W1BQN2 A0A3S2LZG7 H9J0U5 S4PDY3 A0A2H4RMR4 A0A2H4RMU9 D6WK77 A0A0S2Z2X8 K7J584 A0A2J7PEI5 A0A437BEE0 A0A2A4J639 A0A1Q3FJS7 A0A151XFF6 A0A2C9H8Q1 A0A182U9P9 A0A182USL8 B0WIP9 E2C3S6 A0A453YJS9 A0A1S4H0R7 A0A1S4JIR2 A0A158NHN1 A0A212FFX7 A0A182QDV4 A0A2P8Y1N2 E9J2R5 A0A195D4M7 A0A182KCU4 A0A084VM70 A0A2M4BVL7 A0A182M7C8 A0A195AZZ6 A0A2H1WVH3 A0A2M4BVI7 A0A182SLW3 A0A1B6CWG4 A0A088A1P4 U5EUI3 F4WD80 A0A195EF31 A0A0M8ZXJ1 T1DG07 A0A182L6I3 A0A088AQZ8 A0A182JEP7 A0A2M3ZAW7 D6WK78 A0A0L7QVN0 E2B175 A0A195FWL3 A0A1Y1KBD3 W5JFN1 A0A1Y1KBD8 A0A2M4ADQ0 A0A2M4ADP7 A0A182YNG9 A0A194PPJ0 A0A1B6EH81 A0A1Y1MAD0 A0A0C9RS57 A0A1Y1M6Q3 D6WK79 A0A154PSY4 A0A069DR99 A0A182P9H0 E0VDE6 A0A1L8DYZ0 A0A026WVT7 A0A182VUK4
Pubmed
EMBL
BABH01033696
KQ460473
KPJ14454.1
KQ459591
KPI97093.1
MG434596
+ More
ATY51902.1 KZ149932 PZC77317.1 GDQN01011268 JAT79786.1 AGBW02010560 OWR48321.1 ODYU01002128 SOQ39228.1 NWSH01001963 PCG69557.1 GDQN01000489 JAT90565.1 RSAL01000188 RVE44746.1 NWSH01004188 PCG65376.1 AK402126 BAM18748.1 KPI97094.1 MG434606 ATY51912.1 KPJ14453.1 ODYU01001300 SOQ37320.1 KT943565 ALQ33323.1 BABH01033702 BABH01033703 BABH01033704 AGBW02008159 OWR54085.1 JTDY01001677 KOB73179.1 MG434665 ATY51971.1 PCG69556.1 RVE44745.1 KT943561 ALQ33319.1 MG434615 ATY51921.1 BABH01033697 AGBW02011436 OWR46676.1 OWR48320.1 PZC77318.1 RSAL01000098 RVE47627.1 BABH01010352 BABH01010353 GAIX01004762 JAA87798.1 MG434618 ATY51924.1 MG434649 ATY51955.1 KQ971342 EFA03949.1 KT943556 ALQ33314.1 AAZX01000350 NEVH01026096 PNF14739.1 RSAL01000077 RVE48770.1 NWSH01003077 PCG66994.1 GFDL01007164 JAV27881.1 KQ982194 KYQ59109.1 DS231951 EDS28626.1 GL452364 EFN77312.1 APCN01004215 AAAB01008835 ADTU01015717 AGBW02008751 OWR52636.1 AXCN02000844 PYGN01001044 PSN38167.1 GL767998 EFZ12889.1 KQ976870 KYN07796.1 ATLV01014581 KE524975 KFB39064.1 GGFJ01007958 MBW57099.1 AXCM01005439 KQ976694 KYM77615.1 ODYU01010971 SOQ56414.1 GGFJ01007959 MBW57100.1 GEDC01019442 JAS17856.1 GANO01002311 JAB57560.1 GL888086 EGI67807.1 KQ979039 KYN23459.1 KQ435803 KOX73150.1 GAMD01002946 JAA98644.1 GGFM01004847 MBW25598.1 EFA03950.1 KQ414725 KOC62670.1 GL444799 EFN60567.1 KQ981208 KYN44838.1 GEZM01087248 JAV58822.1 ADMH02001356 ETN62876.1 GEZM01087247 JAV58823.1 GGFK01005595 MBW38916.1 GGFK01005593 MBW38914.1 KQ459598 KPI94649.1 GECZ01032471 JAS37298.1 GEZM01038761 JAV81520.1 GBYB01011455 JAG81222.1 GEZM01038762 JAV81519.1 EFA03951.1 KQ435050 KZC14220.1 GBGD01002291 JAC86598.1 DS235073 EEB11402.1 GFDF01002447 JAV11637.1 KK107109 EZA59219.1
ATY51902.1 KZ149932 PZC77317.1 GDQN01011268 JAT79786.1 AGBW02010560 OWR48321.1 ODYU01002128 SOQ39228.1 NWSH01001963 PCG69557.1 GDQN01000489 JAT90565.1 RSAL01000188 RVE44746.1 NWSH01004188 PCG65376.1 AK402126 BAM18748.1 KPI97094.1 MG434606 ATY51912.1 KPJ14453.1 ODYU01001300 SOQ37320.1 KT943565 ALQ33323.1 BABH01033702 BABH01033703 BABH01033704 AGBW02008159 OWR54085.1 JTDY01001677 KOB73179.1 MG434665 ATY51971.1 PCG69556.1 RVE44745.1 KT943561 ALQ33319.1 MG434615 ATY51921.1 BABH01033697 AGBW02011436 OWR46676.1 OWR48320.1 PZC77318.1 RSAL01000098 RVE47627.1 BABH01010352 BABH01010353 GAIX01004762 JAA87798.1 MG434618 ATY51924.1 MG434649 ATY51955.1 KQ971342 EFA03949.1 KT943556 ALQ33314.1 AAZX01000350 NEVH01026096 PNF14739.1 RSAL01000077 RVE48770.1 NWSH01003077 PCG66994.1 GFDL01007164 JAV27881.1 KQ982194 KYQ59109.1 DS231951 EDS28626.1 GL452364 EFN77312.1 APCN01004215 AAAB01008835 ADTU01015717 AGBW02008751 OWR52636.1 AXCN02000844 PYGN01001044 PSN38167.1 GL767998 EFZ12889.1 KQ976870 KYN07796.1 ATLV01014581 KE524975 KFB39064.1 GGFJ01007958 MBW57099.1 AXCM01005439 KQ976694 KYM77615.1 ODYU01010971 SOQ56414.1 GGFJ01007959 MBW57100.1 GEDC01019442 JAS17856.1 GANO01002311 JAB57560.1 GL888086 EGI67807.1 KQ979039 KYN23459.1 KQ435803 KOX73150.1 GAMD01002946 JAA98644.1 GGFM01004847 MBW25598.1 EFA03950.1 KQ414725 KOC62670.1 GL444799 EFN60567.1 KQ981208 KYN44838.1 GEZM01087248 JAV58822.1 ADMH02001356 ETN62876.1 GEZM01087247 JAV58823.1 GGFK01005595 MBW38916.1 GGFK01005593 MBW38914.1 KQ459598 KPI94649.1 GECZ01032471 JAS37298.1 GEZM01038761 JAV81520.1 GBYB01011455 JAG81222.1 GEZM01038762 JAV81519.1 EFA03951.1 KQ435050 KZC14220.1 GBGD01002291 JAC86598.1 DS235073 EEB11402.1 GFDF01002447 JAV11637.1 KK107109 EZA59219.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000283053
+ More
UP000037510 UP000007266 UP000002358 UP000235965 UP000075809 UP000076407 UP000075902 UP000075903 UP000002320 UP000008237 UP000075840 UP000005205 UP000075886 UP000245037 UP000078542 UP000075881 UP000030765 UP000075883 UP000078540 UP000075901 UP000005203 UP000007755 UP000078492 UP000053105 UP000075882 UP000075880 UP000053825 UP000000311 UP000078541 UP000000673 UP000076408 UP000076502 UP000075885 UP000009046 UP000053097 UP000075920
UP000037510 UP000007266 UP000002358 UP000235965 UP000075809 UP000076407 UP000075902 UP000075903 UP000002320 UP000008237 UP000075840 UP000005205 UP000075886 UP000245037 UP000078542 UP000075881 UP000030765 UP000075883 UP000078540 UP000075901 UP000005203 UP000007755 UP000078492 UP000053105 UP000075882 UP000075880 UP000053825 UP000000311 UP000078541 UP000000673 UP000076408 UP000076502 UP000075885 UP000009046 UP000053097 UP000075920
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JSC1
A0A194RAC5
A0A194PVS2
A0A2H4RMP6
A0A2W1BVU1
A0A1E1VYH6
+ More
A0A212F3L1 A0A2H1VEG3 A0A2A4JBT7 A0A1E1WU69 A0A437B2L8 A0A2A4J0W6 I4DLF8 A0A194PWG0 A0A2H4RMQ8 A0A194R9D4 A0A2H1V8Z2 A0A0S2Z3F3 H9JSD0 A0A212FK12 A0A0L7LCR3 A0A2H4RMU7 A0A2A4JCS5 A0A3S2NDL1 A0A0S2Z371 A0A2H4RMQ9 H9JSC0 A0A212EYW0 A0A2W1BQN2 A0A3S2LZG7 H9J0U5 S4PDY3 A0A2H4RMR4 A0A2H4RMU9 D6WK77 A0A0S2Z2X8 K7J584 A0A2J7PEI5 A0A437BEE0 A0A2A4J639 A0A1Q3FJS7 A0A151XFF6 A0A2C9H8Q1 A0A182U9P9 A0A182USL8 B0WIP9 E2C3S6 A0A453YJS9 A0A1S4H0R7 A0A1S4JIR2 A0A158NHN1 A0A212FFX7 A0A182QDV4 A0A2P8Y1N2 E9J2R5 A0A195D4M7 A0A182KCU4 A0A084VM70 A0A2M4BVL7 A0A182M7C8 A0A195AZZ6 A0A2H1WVH3 A0A2M4BVI7 A0A182SLW3 A0A1B6CWG4 A0A088A1P4 U5EUI3 F4WD80 A0A195EF31 A0A0M8ZXJ1 T1DG07 A0A182L6I3 A0A088AQZ8 A0A182JEP7 A0A2M3ZAW7 D6WK78 A0A0L7QVN0 E2B175 A0A195FWL3 A0A1Y1KBD3 W5JFN1 A0A1Y1KBD8 A0A2M4ADQ0 A0A2M4ADP7 A0A182YNG9 A0A194PPJ0 A0A1B6EH81 A0A1Y1MAD0 A0A0C9RS57 A0A1Y1M6Q3 D6WK79 A0A154PSY4 A0A069DR99 A0A182P9H0 E0VDE6 A0A1L8DYZ0 A0A026WVT7 A0A182VUK4
A0A212F3L1 A0A2H1VEG3 A0A2A4JBT7 A0A1E1WU69 A0A437B2L8 A0A2A4J0W6 I4DLF8 A0A194PWG0 A0A2H4RMQ8 A0A194R9D4 A0A2H1V8Z2 A0A0S2Z3F3 H9JSD0 A0A212FK12 A0A0L7LCR3 A0A2H4RMU7 A0A2A4JCS5 A0A3S2NDL1 A0A0S2Z371 A0A2H4RMQ9 H9JSC0 A0A212EYW0 A0A2W1BQN2 A0A3S2LZG7 H9J0U5 S4PDY3 A0A2H4RMR4 A0A2H4RMU9 D6WK77 A0A0S2Z2X8 K7J584 A0A2J7PEI5 A0A437BEE0 A0A2A4J639 A0A1Q3FJS7 A0A151XFF6 A0A2C9H8Q1 A0A182U9P9 A0A182USL8 B0WIP9 E2C3S6 A0A453YJS9 A0A1S4H0R7 A0A1S4JIR2 A0A158NHN1 A0A212FFX7 A0A182QDV4 A0A2P8Y1N2 E9J2R5 A0A195D4M7 A0A182KCU4 A0A084VM70 A0A2M4BVL7 A0A182M7C8 A0A195AZZ6 A0A2H1WVH3 A0A2M4BVI7 A0A182SLW3 A0A1B6CWG4 A0A088A1P4 U5EUI3 F4WD80 A0A195EF31 A0A0M8ZXJ1 T1DG07 A0A182L6I3 A0A088AQZ8 A0A182JEP7 A0A2M3ZAW7 D6WK78 A0A0L7QVN0 E2B175 A0A195FWL3 A0A1Y1KBD3 W5JFN1 A0A1Y1KBD8 A0A2M4ADQ0 A0A2M4ADP7 A0A182YNG9 A0A194PPJ0 A0A1B6EH81 A0A1Y1MAD0 A0A0C9RS57 A0A1Y1M6Q3 D6WK79 A0A154PSY4 A0A069DR99 A0A182P9H0 E0VDE6 A0A1L8DYZ0 A0A026WVT7 A0A182VUK4
PDB
3W68
E-value=1.96528e-29,
Score=320
Ontologies
GO
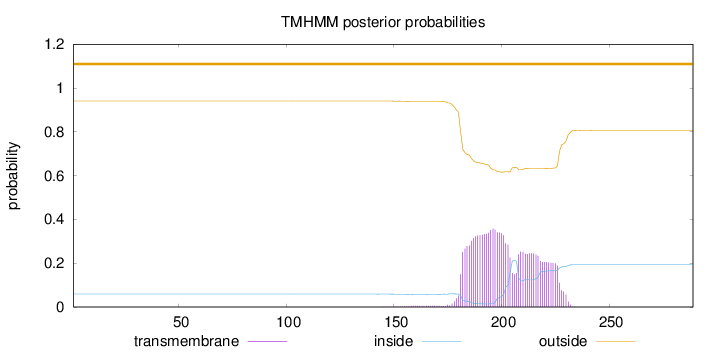
Topology
Length:
289
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.68047
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05908
outside
1 - 289
Population Genetic Test Statistics
Pi
250.448668
Theta
223.783871
Tajima's D
-0.607616
CLR
22.077935
CSRT
0.218339083045848
Interpretation
Uncertain
